Artificial Synthesis of Gene
As mentioned earlier that the molecular structure of yeast alanine tRNA was worked out by R.W. Holley and coworkers in 1965 which helped Khorana to deduce the structure of alanine tRNA. They found out that yeast alanine tRNA contains 77 base pairs. It was very difficult to assemble 77 base pairs of nucleotides in ordered form. Therefore, they synthesized chemically the short deoxynucleotide sequences which was joined by hydrogen bonding to form a long complementary strand. By using polynucleotide ligase the double stranded pieces were produced. The complete procedure of synthesizing gene for yeast alanine tRNA is discussed in the following steps.
(i) Synthesis of Oligonucleotides. In the first approach, fifteen oligonucleotides ranging from pentanucleotide (i.e. oligodeoxynucleotide of five bases) to an icosanucleotide (i.e. oligodeoxynucleotide of twenty bases) were synthesized. The chemical synthesis was brought about through condensation between the - OH group at 3' position of one deoxynucleotide and the - PO4 group at 5' position of the second deoxynucleotide. All other functional groups of deoxyribonucleotides not taking part in condensation processes were protected so that the condensation could be brought about. The deoxynucleotides were protected as below:
(b) The hydroxyl group (-OH) at the 5' position of receiving deoxynucleotide was protected by cyanoethyl group (HN-CH2-CH2-).
(c) The -OH group at the 3' position of second or incoming deoxynucleotide was protected by acetyl group.
The different protecting groups used and treatment required to remove the protecting groups are given in Table 2.4. When the groups of deoxynueleotides were protected, the products reacted to form deoxyoligonucleotide. When deoxynueleotides were condensed into oligonucleotides, different protecting groups were removed by treating with ammonia, acid or alkali (Table 2.5). For example, both the cyanoethyl group at the 5' position and the acetyl group at the 3' position were removed by alkali treatment.
Table 2.5. Different protective groups of nucleotides and their removal
| The group in base or sugar to be protected | Protected by | Protecting group removed by | |
| A. | -NH2 group (base) | ||
| (i) | deoxyadenosine | Benzyl group | Ammonia |
| (ii) | deoxycytidine | Anisoyl group | Ammonia |
| (iii) | deoxyguanosine | Isobutyl group | Ammonia |
| B. | -OH group (sugar) | ||
| (i) | -OH group at 5' position of first nucleotide |
Monomethoxy trityl group | Acid |
| (ii) | -OH group at 5' position of growing chain | Cyanoethyl | Alkali |
| (iii) | -OH group at 3' position | Acetyl group | Alkali |
The joining of the three fragments was done by any of two following methods:
(a) In one approach, fragment A was joined to B by taking advantage of overlapping in nucleotide residues 17-20. Then, the fragment C was added with the overlap in nucleotides 46- 50. Thus, a complete double stranded DNA with 77 base pairs was prepared.
(b) In the second approach, the fragment B was joined to C. At the end the fragment A was added to nucleotide residues 17-20 to obtain the complete gene for alanine tRNA.
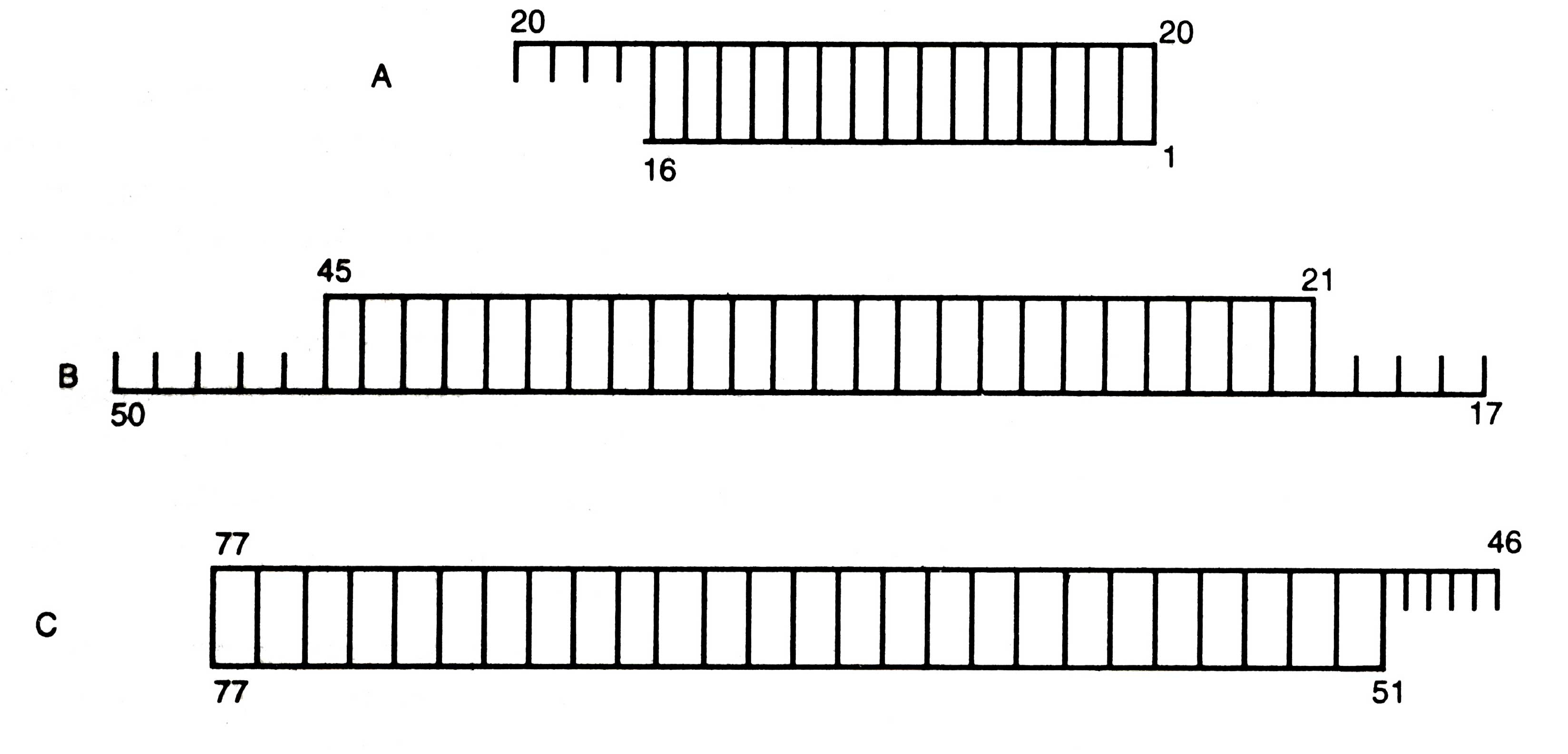
Fig. 2.12. The three duplex DNA fragments (AB, and C) were synthesized for the synthesis of the gene for yeast alanine tRNA
Khorana et. al. (1970) prepared this gene in vitro which was used for future work. They found that alanine tRNA gene replicated and transcribed into tRNA just like the natural gene. It is not known whether tRNA prepared from artificially synthesized gene had the molecular organization similar to alanine tRNA or not.
Artificial Synthesis of a Gene for Bacterial tyrosine tRNA
In 1975, Khorana and co-workers completed the synthesis of a gene for E. coli tyrosine tRNA precursor. E. coli tRNA precursors are formed from the larger precursors. The tyrosine tRNA precursor has 126 nucleotides. They synthesized the complete sequence of DNA duplex coding for tyrosine tRNA precursor of E. coli, and promoters are terminator genes. Though these segments are not the proper structural gene yet are the regions involved in its regulation.
Khorana (1979) completely synthesized a biologically functional tyrosine tRNA suppressor gene of E. coli which was 207 base pairs long and contained (i) a 51 base pairs long DNA corresponding to promoter region, (ii) a 126 base pair long DNA corresponding to precursor region of tRNA, (iii) a 25 base pair long DNA including 16 base pairs contained restriction site for EcoRI. This complete synthetic gene was joined in phage lambda vector which in turn was allowed to transfect E. coli cells. After transfection phage containing synthetic gene was successfully multiplied in E. coli.
Khorana (1979) made the phosphodiester approach for synthesizing the oligonucleotides of the biologically active tRNA. The demerits of this approach are: (i) the completion of reaction in long time, (ii) rapidly decrease in yield with the increase in chain length, and (iii) time taking procedure of purification.
Interferons are proteinaceous in nature produced in human to inhibit viral infection. These are of three types secreted by three genes i.e. (i) leukocyte interferon gene (IFN-oc gene), (ii) fibroblast interferon gene (IFN-p gene), and (iii) immune interferon gene (IFN-γ gene). In 1980, Weismann and co-workers published the nucleotide sequence of IFN-a gene. Taking advantage of this information Edge et al. (1981) successfully synthesized the total human interferon gene of 514 base pairs long.
Edge et al. (1981) made the phosphotriester approach in artificially synthesizing 67 oligonucleotides of 10-20 nucleotide residue long segment. The phosphotriester approach overcomes some of the demerits of phosphodiester approach by blocking the function of each internucleotide phosphodiester during the process of synthesis. A completely protected mononucleotide containing a fully masked 3' phosphate triester group is used in this method.
Edge et. al. (1981) incorporated the artificially synthesized gene into a plasmid through biotechnological technique (see Tools of Genetic Engineering and Techniques of Genetic Engineering). The recombinant plasmid was transferred into E.coli cells which expressed oc-interferon. This technique now-a-days is being adopted to produce interferon commercially.




