Nucleic Acid Detection Assays
Genes contain the genetic message (genotype) for all forms of life. The expression of the genotype results in the production of physical characteristics (phenotype) that make each life form special and unique. All genes are made up of nucleic acids consisting of either DNA or RNA. In recent years, molecular biologists have been able to determine the order, or sequence, in which the nucleotides adenine, thymine, cytosine, and guanine (or uracil in RNA) occur in these nucleic acid molecules. Sequencing has revealed the entire set of genes (i.e., the genome) of many life-forms including various microorganisms, and even humans. By comparing the nucleotide sequence of genes among various life-forms, scientists can determine common regions of nucleic acids but, more importantly, they can determine the different nucleotide sequences that make a life-form special and unique.With this knowledge, and the understanding that the nucleotide base adenine always bonds to thymine (in DNA) or uracil (in RNA), and guanine always bonds to cytosine, it is possible to synthesize in the laboratory a single-stranded sequence of nucleotides, known as a primer, that is complementary to a unique gene sequence in a specific life-form. When two single nucleic acid strands with complementary base sequences are placed together in solution, the nucleotide base pairs of each strand bond together to form a double-stranded molecule, called a duplex or hybrid. This hybridization reaction serves as the basis of two nucleic acid detection methods: probe assays and amplification assays.
Like the antigen detection assays just described, these nucleic acid detection assays have come into common use in many clinical microbiology laboratories, either to confirm the identity of a microorganism or to detect its presence directly in a clinical sample.
The basic principles of probe and amplification assays are reviewed here.
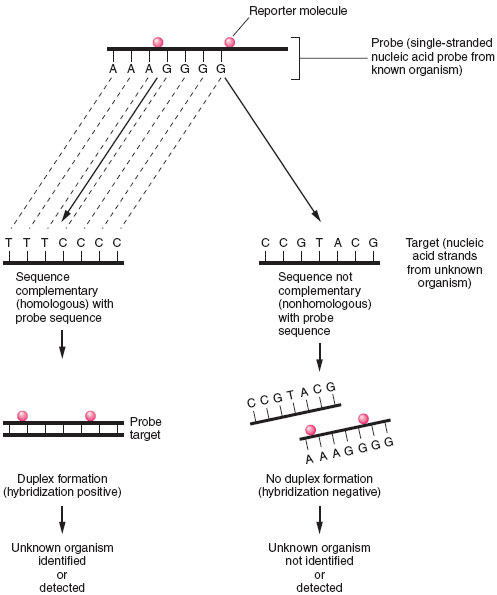 |
| Figure 19.5 Principles of nucleic acid hybridization. Identification of unknown organism is established by positive hybridization (i.e., duplex formation) between a probe nucleic acid strand (from known organism) and a target nucleic acid strand from the organism to be identified. Failure to hybridize indicates lack of homology between probe and target nucleicacid. |
Probe Assays
In a probe hybridization assay, one nucleic acid strand, known as the probe, will seek a complementary nucleic acid strand, the target, with which to combine. The probe is derived from a known microorganism and the target is an unknown microorganism present in a clinical sample or isolated in culture. Like antigen detection assays, a marker or reporter molecule must be attached to the probe to determine whether the hybridization reaction has taken place (see fig. 19.5). Among the more popular reporter molecules are 32P or 125I; enzyme conjugates of alkaline phosphatase or horseradish peroxidase; and chemiluminescent molecules, such as acridinium. Chemiluminescent reporter molecules emit light that can be measuredby using a special instrument called a luminometer. The amount of reporter molecule detected is directly proportional to the amount of hybridization that has occurred.
A positive hybridization reaction indicates that the unknown target is the same as the organism that served as the probe source. If no hybridization is detected, the target organism is not present in the sample (see fig. 19.5).
Single-stranded probe molecules can be composed of either DNA or RNA. Thus, double-stranded hybrids may be DNA-DNA, RNA-RNA, or DNA-RNA. Commercially available kits in which the probe molecule is DNA and the target molecule is a complementary strand of ribosomal RNA are popular because a microbial cell has several thousand copies of ribosomal RNA sequences but only one or two copies of DNA targets. As a result, these RNA-directed probes are far more sensitive for detecting low numbers of a microorganism in a sample than are DNA-directed probes.
Probe hybridization assays are used to confirm the identity of a wide variety of bacteria, fungi, viruses, and protozoa. Although they were once used commonly to detect the sexually transmitted bacterial pathogens, Chlamydia trachomatis and Neisseria gonorrhoeae, directly in clinical specimens, the more sensitive amplification assays have gained popularity instead.
Amplification Assays
Often, too few bacteria may be present in a clinical specimen to be detected by a probe assay. To overcome this problem, a variety of methods referred to as amplification techniques have been devised. The most popular of these (for which Dr. Kary Mullis won the Nobel prize) is called the polymerase chain reaction or PCR. In this method, the specimen is heated to separate bacterial DNA strands, known bacterial primers are added to the mixture, and if they match the unknown single-stranded DNA, they combine (anneal) with it. Because the primers are shorter sequences than the original DNA strands, nucleotides and a heatresistant Taq polymerase enzyme (originally isolated from a bacterium living in a hot spring in Wyoming’s Yellowstone National Park) are added to the mixture to complete the formation of the double-stranded DNA. The new double-stranded DNA, referred to as an amplicon, is again separated, annealed with new primers, and extended with nucleotides in the presence of the polymerase enzyme. Each PCR step is carried out at a different temperature, which is automatically controlled by an instrument known as a thermocycler. The cycling is continued for up to 40 cycles, during which the original DNA sequences are increased or amplified a billionfold and, thus, many copies are available for detection. Probes labeled with reporter molecules that provide a chemiluminescent or EIA-type colored signal are popular methods for amplicon detection. Figure 19.6 illustrates the steps in the PCR reaction.
Other nucleic acid amplification assays have been developed for the rapid detection of microorganisms in clinical specimens, but their principle is similar to that of PCR. An amplification assay for detecting any infectious agent can be developed as long as the appropriate primer sequence is available to begin the amplification reaction. Because these assays are so sensitive, great care must be taken in the laboratory to avoid contaminating clinical samples with extraneous DNA that could be amplified and result in false-positive reactions.
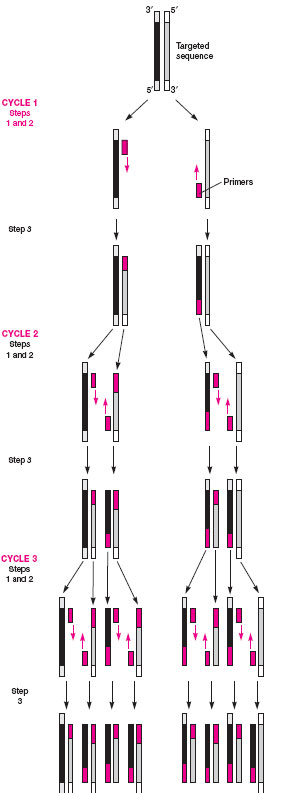 |
| Figure 19.6 Three cycles of a PCR reaction: after 40 cycles, the DNA sequences are amplified a billionfold. |




