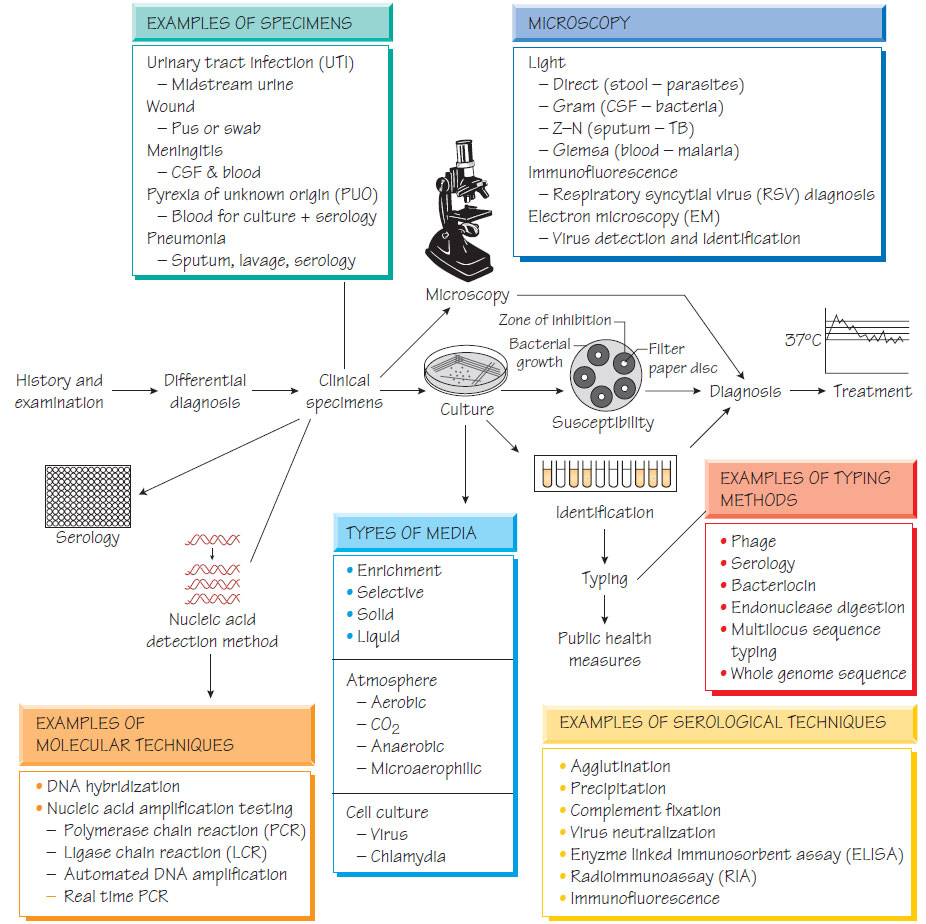
The laboratory investigation of infection
SpecimensAny tissue or body fluid can be subjected to microbiological investigation with the aim of identifying the infecting pathogen and predicting response to therapy.
To optimize the diagnostic benefit it is necessary to:
- understand in which tissues/specimens the organism is to be found and when in the natural history of the infection;
- take samples carefully (e.g. poor aseptic technique may lead to contamination of sterile samples causing false-positive results);
- transport samples rapidly to the laboratory in a suitable medium.
It is important to remember that some organisms survive poorly outside the body (e.g. numbers of strict anaerobes are reduced by atmospheric oxygen) and in some cases suitable media needs to be inoculated directly in the clinic (e.g. for isolation of Neisseria gonorrhoeae).
Specimens may be examined grossly, for example to detect adult worms in faeces. Although microscopy is rapid, it is insensitive and requires considerable expertise; specificity may also be a problem if commensal organisms can be mistaken for pathogens. Microscopy can also be used to define specimen quality, for example identifying salivary contamination of sputum by the presence of epithelial cells.
Special stains can be used to help identify organisms, such as Giemsa staining of blood films and tissues, which is used to identify malaria and Leishmania (Malaria, leishmaniasis and trypanosomiasis). Immunofluorescence can provide precise identification of a pathogen by using antibodies that are directed against a specific organism.
- Antibiotic therapy administered presampling can falsely render samples culture negative.
- Most human pathogens are fastidious, requiring media supplemented with nutrients to support growth and increase their numbers to detectable levels.
- Growth on solid media allows organisms to be separated into individual colonies; a pure (clonal) population permits subsequent identification and susceptibility testing.
- Selective agents such as antibiotics or dyes may be used to suppress unwanted organisms in specimens with a normal flora.
- An appropriate atmosphere must be provided: fastidious anaerobes require an oxygen-free atmosphere.
- Most pathogenic bacteria are incubated at 37 °C, but some fungi are incubated at 30 °C.
- Identification predicts pathogenicity: Vibrio cholerae causes severe watery diarrhoea, whereas Shigella sonnei infection is usually mild.
- Identification of some organisms should prompt public health action, for example contact tracing for a patient found to have meningococcal meningitis.
Bacterial identification depends on colonial morphology on agar, microscopic morphology, biochemical tests and, increasingly, nucleic acid amplification tests (NAATs) and gene sequencing. This is especially important for organisms that are slow growing (e.g. Mycobacterium tuberculosis) or impossible to grow (e.g. Trophyrema whippelii) to grow.
Susceptibility testing aims to determine whether treatment with a given antibiotic will be successful. A susceptible organism should respond to a standard dose of an antimicrobial, a moderately resistant strain should respond to a larger dose, whereas a resistant organism is likely to fail therapy with the given antibiotic. Clinical response depends on host factors, and in vitro tests only provide an approximate guide to therapy.
Several bodies including the British Society of Antimicrobial Chemotherapy (BSAC) and the Clinical Laboratory Standards Institute (CLSI) define methods and standardized conditions to ensure testing is reproducible. Both are based on measurement of the diameter of the zone of inhibition of confluent growth for the test organism that is caused by an antimicrobial incorporated into a paper disc. The minimum inhibitory concentration, which is the lowest dose that completely inhibits growth, is a more objective method and enables resistance levels to be related to the concentration of antibiotic that is achievable in the tissues.
Susceptibility can be assessed rapidly by hybridization or sequence-based methods that detect specific antibiotic-resistance mutations.
An infection can be diagnosed by detecting the immune response to the pathogen: for example by detection of rising or falling antibody concentrations more than a week apart, or by the presence of a specific IgM or specific antigen. These techniques are used for organisms that are difficult to grow such as viruses (e.g. HIV or hepatitis B).
Molecular techniques
Southern blotting and nucleic acid hybridization
A labelled DNA probe will bind to the specimen if it contains the specific sequence that is being sought. The captured probe is detected by the activity of it attached label. This technique is specific and rapid, but less sensitive than other methods that involve amplification steps.
Nucleic acid amplification tests (NAATs) make the diagnosis by amplifying specific regions of the genome from the pathogen. Although different methods are used to amplify pathogen-specific DNA or RNA the aim is the same, to produce sufficient copies for detection. For example, nucleic acid from the pathogen is separated into single strands and primers are designed to bind to target sequences. A polymerase then catalyses synthesis of new nucleic acid and this process is repeated for multiple cycles. Automated systems and commercial kits have made these tests available in many laboratories. Real-time PCR machines measure rising concentrations of target DNA and determine positivity when the concentration passes a set threshold. NAATs have the advantage that they can detect slow-growing organisms or those that are difficult to grow (e.g. M. tuberculosis) or make a diagnosis when samples are rendered falsely negative by antibiotic therapy. Methods to detect antibiotic-resistance genes can also be used to provide surrogate susceptibility results (e.g. detection of the rpoB gene mutation for rifampicin resistance in M. tuberculosis).