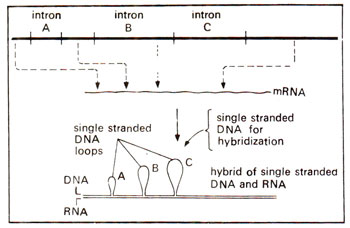
Fig. 29.1. The upper figure shows a DNA sequence representing an interrupted gene with their introns (A, B, C), and the synthesis of its mRNA; the bottom figure shows the result of hybridization of mRNA with single stranded DNA obtained after denaturation of native DNA (note the loops formed by three intron regions).
After rediscovery of Mendel's laws in 1900, a gene was considered earlier to be a point on a chromosome, and later to be a continuous segment of DNA. It was also proved as late as 1964 that there was co-linearity between sequence of nucleotides on DNA and sequence of amino acids in corresponding protein (consult
Mutations: 3. Molecular Level (Mechanism)), so that there was absolutely no doubt about the continuity of nucleotides in a gene represented by a DNA segment. This was proved to be true not only in prokaryotes but also in eukaryotes. In view of this, it was a big surprise when, in 1977,
it was proved for the first time that in some mammals, birds and amphibians, a gene may not be represented by a continuous sequence of nucleotides but may be interrupted by some intervening sequences which are not represented in mRNA transcribed from the gene and utilized for the synthesis of proteins. Such genes with intervening sequences were called split genes (Fig. 29.1).

Fig. 29.1. The upper figure shows a DNA sequence representing an interrupted gene with their introns (A, B, C), and the synthesis of its mRNA; the bottom figure shows the result of hybridization of mRNA with single stranded DNA obtained after denaturation of native DNA (note the loops formed by three intron regions).
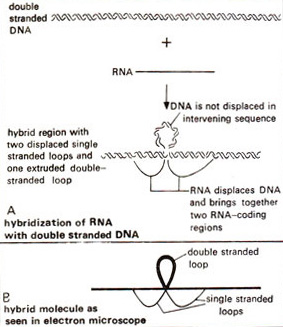
Fig. 29.3. R-looping, where mRNA is hybridized with double stranded DNA (note the double stranded thick loop showing intron region and single stranded thin loops representing the exons).
The discovery of split genes was made in 1977 by several groups in a variety of materials, which included the following : (i) Two research groups separately headed by
Phillip A. Sharp and
Richard J. Roberts studied genes of adenovirus 2. (ii) Research groups of
D.S. Hogness, I.B. David and
N. Davidson studied genes for 28S rRNA in
Drosophila. (iii) Research groups of
P. Chambon, P. Leder and
R.A. Flavell studied β globin genes, ovalbumin genes and tRNA genes. In all these cases the genes were found to be interrupted by intervening sequences. The credit for discovery of split genes, however, goes to Phillip Sharp and Richard Roberts,
who won the 1993 Nobel Prize for Medicine for their work on split genes. They analysed the hybrids of late mRNA of adenovirus 2 with the adenovirus genomic DNA. When these mRNA-DNA hybrids were examined under electron microscope, the adjoining sequences of mRNA were found to hybridize with discontinuous stretches of genomic DNA of adenovirus 2. The intervening DNA sequences were observed as loops and the phenomenon was later described as R-looping (see later for R-looping, see Fig. 29.3). Chambon's group compiled sequences of the boundaries of introns from a large number of protein coding eukaryotic genes (not ribosomal RNA or tRNA genes), which revealed the presence of consensus sequences at the intron-exon junctions. Of these GT was always found at the 5' side of the intron (left splice junction) and AG at the 3' side (right splice junction). This became popularly known as GT-AG or Chambon's rule. Some of the diseases (e.g. thalassemias) are caused by mutations, which created or abolished these splice junctions. Phillip Sharp's group and other groups later also conducted studies to elucidate the mechanisms of splicing (for details, consult
Expression of Gene : Protein Synthesis 3. RNA Processing (RNA Splicing, RNA Editing and Ribozymes)).

Fig. 29.3. R-looping, where mRNA is hybridized with double stranded DNA (note the double stranded thick loop showing intron region and single stranded thin loops representing the exons).
A detailed study was conducted on
ovalbumin gene found in chicken. This
ovalbumin gene is responsible for synthesis of a protein 'ovalbumin' consisting of 386 amino aicds and synthesized only by highly specialized tubular gland cells of the oviduct at the time when the hen is laying eggs. The expression of this
ovalbumin gene is controlled by some female sex hormones.
Pierre Chambon and his colleagues synthesized artificial
ovalbumin gene in order to study its regulation. Such an artificial gene could be synthesized by using
ovalbumin mRNA which could give rise to cDNA (
complementary DNA)with the help of enzyme reverse transciptase. This cDNA was inserted into a plasmid and cloned in
E. coli for its multiplication (consult
Genetic Engineering and Biotechnology 1. Recombinant DNA and PCR (Cloning and Amplification of DNA)). When this cDNA was compared with corresponding genomic DNA, it was discovered (through DNA hybridization) that the genomic DNA had additional intervening sequences.
Another important technique utilized for the study of split genes was the use of restriction enzymes which have the property of cleaving DNA at unique sites. More than 100 such restriction endonuclease enzymes are now available (consult
Chemistry of the Gene 2. Synthesis, Modification and Repair of DNA and
Genetic Engineering and Biotechnology 1. Recombinant DNA and PCR (Cloning and Amplification of DNA)). In view of this, restriction endonucleases could by used to find out the presence or absence of a certain sequence in a particular gene. For example, when
EcoR1and
HindIIIenzymes were used with cDNA for ovalbumin, it was found that no cleavage occurred suggesting that the sequences of six base pairs each recognized by these two enzymes were absent.
On the basis of this it was expected that if the DNA extracted from oviduct or another tissue was cleaved by utilizing these two enzymes, the
ovalbumin gene will not be broken and the cleavage will occur at other places thus making it possible to isolate the native
ovalbumin gene from the living cells. This
: DNA segment representing
ovalbumin gene was expected to be separated with the help of hybridization with cDNA artificially synthesized. When such hybridization was done, cDNA hybridized with different fragments of DNA rather than with a single fragment. Hybridization between the single stranded DNA having the gene for ovalbumin and its mRNA also showed formation of distinct loops at specific sites as observed in electron microscope (Fig. 29.1).
This DNA forming the loops is obviously absent in mRNA. This kind of results eventually led to the confirmation of the presence of split genes in 1977. Subsequently it could be proved that split genes are present atleast in two more cases i.e. gene for β
globin (a component of haemoglobin molecule) in rabbit and mouse and
immunoglobulin gene (antibody gene). By the end of 1977, it was fairly clear that .the presence of split genes in case of higher organisms was a phenomenon of general occurrence.
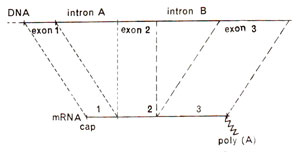
Fig. 29.2. Diagrammatic representation of a hypothetical split gene having three exons (exon 1, exon 2, exon 3) and two introns (intron A, intron B) and its relationship with mRNA. There are no sequences in mRNA corresponding to those in two introns (redrawn from Science. Vol. 204, 1979).
We may discuss the problem of split genes by taking a hypothetical example of a gene represented by a DNA sequence which has three pieces, called exon 1, exon 2 and exon 3 (Fig. 29.2). These three exons are separated by two long intervening sequences called introns A and B. The terms exon and intron were used by
W. Gilbert for the first time and are being followed ever since. Figure 29.2 shows,
that in the transcription of DNA, the ultimate product i.e. mRNA had only those sequences which corresponded to exons, the sequences representing introns being entirely absent. The mechanism of production of mRNA from DNA sequences having intervening sequences called introns was a matter of discussion during 1978 and 1979. Later it was discovered that both exons and introns are first transcribed and that this primary transcript is then modified. The sequences corresponding to introns are removed and the sequences corresponding to exons are joined together, in correct order to give rise to mRNA (consult
Expression of Gene : Protein Synthesis 3. RNA Processing (RNA Splicing, RNA Editing and Ribozymes) for more details). A generalization has also been made that the order of exons on DNA is the same as the order in which they are found in processed mRNA. There does not seem any strong reason why it should always be true and it is possible that the order of exons would sometimes be different.

Fig. 29.2. Diagrammatic representation of a hypothetical split gene having three exons (exon 1, exon 2, exon 3) and two introns (intron A, intron B) and its relationship with mRNA. There are no sequences in mRNA corresponding to those in two introns (redrawn from Science. Vol. 204, 1979).
It has also been shown that all classes of genes can be interrupted. These interrupted genes may include (i) nuclear genes for proteins, (ii) nuclear genes coding for rRNA, (iij) nuclear genes coding for tRNA, (iv) mjtochondrial genes in yeast, (v) chloroplast genes in a wide variety of plants, (vi) genes in archaebacteria and (vii) genes in bacteriophages of
E. coli. However, they are entirely absent in eubacterial genomes. (Recently f introns were discovered even in eubacteria).










