In Situ Hybridization Applicable to mRNA Species in Cultured Cells
I. INTRODUCTIONRNA in situ hybridization (ISH) techniques allow the study of gene expression at the individual cell level in a histocytomorphological context. These techniques often involved the use of radioactive-labeled probes. However, because of resolution drawbacks limiting their applicability in the study of RNA distribution at (sub)cellular sites, these probes are replaced more frequently by hapten- or fluorophore-labeled ones. Nevertheless, radioactive ISH is still useful for detecting relatively low abundant RNA transcripts in tissue sections.
From the early 1980s, several nonisotopic-labeling methods have been developed that are most often based on the introduction of haptens (e.g., biotin and digoxigenin) or fluorochromes (e.g., fluorescein, rhodamine, coumarin, Cy3, and Cy5) that are conjugated to allyl or alkylamine-dUTP in DNA or RNA probes. Such probes provide a high spatial resolution and allow simultaneous detection of multiple RNA sequences (Dirks et al., 1991; Levsky et al., 2002) or RNA sequences together with proteins (Dirks, 1998; Snaar et al., 1999).
Direct ISH techniques, using fluorochrome-labeled probes, are used to detect relatively abundant mRNA species by conventional fluorescence microscopy. Using more advanced digital imaging microscopy, visualization of single RNA transcripts proved even possible when oligodeoxynucleotide probes were labeled with five fluorochromes per molecule and 10 oligonucleotide probes were hybridized to adjacent sequences on an RNA target (Femino et al., 1998).
Indirect ISH techniques, using haptenized probes, are often the method of choice because of their better sensitivity compared to direct fluorescence ISH approaches (Dirks et al., 1993). Furthermore, enhancement of ISH signals can be accomplished by applying multiple antibody layers or by the use of a tyramidebased detection method (Adams, 1992; Raap et al., 1995; van de Corput et al., 1998). Tyramide-based detection methods involve the use of an antihapten peroxidase-labeled antibody as a first antibody layer and a biotin, DNP, or fluorochrome tyramide that serves as a peroxidase substrate to generate and deposit many reporter molecules close to the hybridization site.
This article describes a protocol that allows sensitive detection of RNAs at cytoplasmic as well as at nuclear sites of cells that are grown or attached to microscopic slides.
II. MATERIALS AND INSTRUMENTATION
Dulbecco's modified Eagle medium without phenol red (DMEM, Cat. No. 1188-36), fetal bovine serum (FBS, Cat. No. 1050-64), l-glutamine (Cat. No. 2503-24), penicillin-streptomycin (Cat. No. 1514-22), and trypsin (Cat. No. 2509-28) are from Invitrogen Life Technologies. Salmon testes DNA (Cat. No. D-7656), dithiothreitol (DTT, Cat. No. D-0632), thimerosal (Cat. No. T-5125), polyvinylpyrrolidone (PVP, Cat. No. PVP- 40), Tween 20 (Cat. No. P-1379), mouse monoclonal antidigoxin (Cat. No. D-8156), rabbit antimouse fluorescein isothiocyanate (FITC) (Cat. No. F-7506), Streptavidin- FITC (Cat. No. S-3762), and goat antirabbit FITC (Cat. No. F-9262) are from Sigma. Ficoll PM 400 (Cat. No. 1-30-0) is from Amersham Biosciences. Bovine serum albumin fraction V (BSA, Cat. No. 44155) is from BDH. Formamide (Cat. No. 7042) and acetic acid (Cat. No. 6052) are from J. T. Baker. Amberlite MB1 ion exchanger (Cat. No. 40701) and 4,6,- diamidino-2-phenylindol 2HCl (DAPI, Cat. No. 18860) are from Serva. Acid-free formaldehyde (Cat. No. 3999) is from Merck. dATP (Cat. No. 1051 440), dCTP (Cat. No. 1051 458), dGTP (Cat. No. 1051 466), dTTP (Cat. No. 1051 482), DNase I (Cat. No. 104 158), digoxigenin-11-dUTP (Cat. No. 1093 088), sheep antidigoxigenin HRP (Cat. No. 1207 733), and blocking reagent (Cat. No. 1096 176) are from Roche. DNA polymerase I (Cat. No. M2051) is from Promega. Vectashield mounting medium is from Vector. Staining jars (100ml) for object slides (Cat. No. L4110) are from Agar Aids Ltd. TSA fluorescein system (Cat. No. NEL701), TSA tetramethylrhodamine NEL702), TSA coumarin system (Cat. No. NEL703), and TSA biotin system (Cat. No. NEL700) are from NEN.
Fluorescence ISH results were examined with an epifluorescence microscope (DM, Leica) equipped with a 100-W mercury arc lamp and a triple excitation filter for red, green, and blue excitation (Omega). Digital images were captured with a cooled CCD camera (Photometrics).
III. PROCEDURES
A. Labeling of DNA by Nick Translation
Solutions
- Nick translation buffer (10x): 0.5M Tris-HCl, pH 7.8, 50mM MgCl2, and 0.5mg/ml BSA. To make 10ml of the solution, add 5ml of an autoclaved 1M stock solution of Tris-HCl, pH 7.8, 0.5 ml of an autoclaved 1M solution MgCl2, and 5 mg of nuclease-free BSA and complete to 10ml with autoclaved distilled water. Aliquot in 100-µl portions and store at -20°C.
- 0.1M DTT: To make 10ml, dissolve 150mg of DTT in 10ml of 0.01M sodium acetate (pH 5.2). Filter the solution through a 0.2-µm filter, aliquot in 1-ml portions, and store at -20°C.
- Nucleotide mix: 0.5mM dATP, 0.5mM dCTP, 0.5mM dGTP, and 0.1mM dTTP. To make 1ml of the solution, add 5µl each of 100mM stock solutions dATP, dCTP, and dGTP and 1µl of 100mM stock solution dTTP. Complete to 1ml with autoclaved distilled water and store at -20°C.
- 1mg/ml DNase I: To make 1ml, add 1mg of DNase, 20µl of a 1M stock solution of Tris-HCl, pH 7.6, 50µl of a stock solution of NaCl, 10µl of a 100mM stock solution of DTT, 0.1mg of BSA, and 0.5ml of glycerol. Complete to 1ml with autoclaved distilled water and store at -20°C.
- 10mg/ml salmon testes DNA: To make 10ml, dissolve 100mg DNAin 100ml 0.3M NaOH in TE, pH 7.8 (10mM Tris-HCl, pH 7.8, 1mM EDTA). Boil for 20 min at 100°C. Neutralize the solution by adding 5ml of 2M Tris-HCl, pH 7.5, 7.5ml of 4M HCl, and 12ml of 2M sodium acetate. Precipitate the DNA by adding 2 volumes of 100% -20°C ethanol and incubate for 1h on ice. Centrifuge for 10min at 2600g, remove the ethanol, and dissolve the DNA pellet in 10 ml TE buffer.
- Deionized formamide: To make 100ml, add 5g of ion exchanger to 100ml formamide. Stir for 2h and filter twice through Whatmann No. 1 filter paper. Aliquot in 1-ml portions and store at -20°C.
- 20x SSC: To make 1 liter, add 175.3g NaCl and 88.24 g sodium citrate to distilled water. Adjust to pH 7.0 and complete to 1 liter. Autoclave the solution and store at room temperature.
- 50X Denhardt's solution: 1% PVP, 1% Ficoll (type 400), and 1% BSA. To make 500ml of the solution, add 5g Ficoll, 5g PVP, 5g BSA (fraction V), and distilled water to 500ml. Sterilize the solution by filtration and store at -20°C.
Steps
- Thaw the required stock solutions and keep them on ice. Prepare the labeling solution on ice and mix well. To make the labeling mixture, combine 26µl autoclaved distilled water, 5µl 10x nick translation buffer, 5 µl 100mM DTT, 4 µl nucleotide mix, 2 µl digoxigenin- 11-dUTP (0.25mM stock solution), 1µl probe DNA (1µg, e.g., cDNA), 2µl DNA polymerase I, and 5µl of a 1:1000 DNase I dilution from a 1-mg/ml stock solution.
- Place the labeling mixture for 2 h in a 16°C water bath.
- Add 250µl distilled water and precipitate the labeled probe by adding 30µl of 3M sodium acetate, pH 4.8, 5 µl salmon testes DNA, and 750µl -20°C 100% ethanol. Mix well and place the solution on ice for 30 min.
- Centrifuge for 30 min at 4°C in an Eppendorf centrifuge at maximum speed. Remove the ethanol completely and resuspend the pellet in 200µl hybridization mixture to reach a probe concentration of 5ng/µl. To make 10ml hybridization mixture, combine 5ml deionized formamide, 1ml 20X SSC, 1ml of 0.5M sodium phosphate buffer, pH 7.0, 1ml 50x Denhardt's solution, and 2ml autoclaved distilled water. The probe mixture can be stored at 4°C for at least 1 year.
B. Culturing and Fixation of Cells
Solution
10x PBS (pH 7.2): Add 80g NaCl, 2g KCl, 15g Na2HPO4·2H2O, and 1.2 g KH2PO4 and adjust to 1 liter. Autoclave the solution and store at room temperature.
Steps
- Trypsinize subconfluent cells that are grown in tissue culture flasks as a monolayer in DMEM supplemented with 10% FCS, 100 U of penicillin/ml, and 100µg of streptomycin/ml and seed into petri dishes containing five microscopic object slides and 20ml medium. Grow cells to subconfluency at 37°C in a humidified 5% CO2 atmosphere.
- Wash the object slides containing cells in PBS for I min at room temperature.
- Fix the cells in PBS containing 3.7% formaldehyde, 5% acetic acid for 20min at room temperature. To make 100ml fixative, combine 75 ml distilled water, 10 ml 10x PBS, 10 ml formaldehyde (37% stock), and 5 ml acetic acid.
- Wash the cells in PBS for 5 min at room temperature and transfer the object slides to a 100-ml staining jar.
- Wash the cells in 70% ethanol and store the object slides in 70% ethanol at 4°C until use.
C. Pretreatment and Hybridization
Solution
0.1% pepsin: To make 100ml, dissolve 0.1 g pepsin in distilled water, adjust pH to 2.0, and bring to 100ml. Make this solution about 15 min before use and place in a 37°C water bath.
Steps
- Wash the cells in PBS for 3 min at room temperature.
- Incubate the cells in 0.1% pepsin solution for 2min at 37°C.
- Wash the cells in 70% ethanol for 30s.
- Wash twice in PBS for 30s each.
- Incubate in 1% formaldehyde in PBS for 5 min at room temperature.
- Wash in PBS for 5 min and dehydrate the cells successively in 70, 90, and 100% ethanol for 3 min each.
- Denature the probe dissolved in hybridization mixture for 5 min in an 80°C water bath.
- Place the probe for I min on ice and spin down in a table centrifuge.
- Apply 10 gl of the denatured probe mixture on the object slide and hybridize overnight at 37°C in a moist chamber, which consists of a l-liter beaker covered with aluminium foil containing tissues moistened with 50% formamide/2x SSC, pH 7.0.
D. Posthybridization Washes Solutions
- 50% formamide/2x SSC wash solution: To make 400ml, add 200ml formamide, 40ml 20x SSC, and 160ml distilled water and adjust the pH to 7.0.
- 10x TBS: To make 1 liter, dissolve 121.4g Tris and 87.4 g NaCl in 800ml distilled water. Adjust the pH to 7.4 with 6 N HCl and bring to a total volume of 1 liter.
Steps
- Wash the slides in the 50% formamide/2x SSC solution at room temperature until the coverslips are released.
- Transfer the slides to a new jar filled with 100ml 50% formamide/2x SCC and wash for 10min.
- Wash the slides for 10min each in 50% formamide/ 2x SSC at 42°C and in the same solution at room temperature.
- Wash the slides in 2x SSC for 5min at room temperature.
- Wash the slides in 1× TBS for 5min at room temperature.
E. Conventional Immunocytochemical Detection
Solution
Blocking solution: To make 100ml, add 0.5 g blocking reagent and 100µl thimerosal (to prevent bacterial growth). Complete to 100ml with 1× TBS. Heat the mixture for 1h at 60°C to dissolve the blocking reagent and aliquot in 10-ml portions. Store at -20°C.
Steps
- Take a slide from the TBS solution and apply 100µl blocking solution. Cover it with a 24 × 50-mm2 coverslip and incubate for 30 min at 37°C in a moist chamber (l-liter beaker covered with aluminium foil containing tissue moistened with water).
- Wash briefly with TBS to remove the coverslips.
- Drain of as much of the TBS solution as possible and incubate the slides with mouse antidigoxigenin, diluted 1:500 in blocking solution under a coverslip for 45min at 37°C in a moist chamber.
- Remove the coverslips by a brief wash in TBS and wash the slides 3 × 5min with TBS at room temperature.
- Drain of as much of the washing solution as possible and incubate the slides with rabbit antimouse FITC diluted 1:500 in blocking solution as described in step 3.
- Wash the slides as described in step 4.
- Drain of as much of the washing solution as possible and incubate the slides with goat anti-rabbit FITC, diluted 1:500 in blocking solution, as described in step 3.
- Wash the slides as described in step 4.
- Dehydrate the slides for 3 min each in 70, 90, and 100% ethanol and air dry.
- Apply 30µl Vectashield containing 10ng/µl DAPI (blue fluorescent DNA counterstain) on a slide and cover with a 24 × 50-mm2 coverslip.
- Examine the slides with a fluorescence microscope equipped with appropriate excitation and emission filters for FITC and DAPI fluorescence.
F. Tyramide Signal Amplification
Solution
10x TNT: To make 1 liter, dissolve 121.4g Tris and 87.4 g NaCl in 800ml distilled water. Ad 0.5 ml Tween 20. Mix thoroughly, adjust the pH to 7.4, and bring to a total volume of 1 liter.
Steps
- Take slides from the TBS solution and apply 100µl blocking solution. Cover with a coverslip and incubate for 30min at 37°C in a moist chamber.
- Wash briefly with TBS to remove coverslips.
- Drain of as much of the washing solution as possible and incubate the slides with antidigoxigenin- HRP, diluted 1:500 in blocking solution, under a coverslip for 45 min at 37°C in a moist chamber.
- Remove the coverslips by a brief wash in TNT and wash the slides 3 × 5min with TNT at room temperature.
- Drain of as much of the TNT solution as possible and apply 300µl of a 1:50 dilution of a fluorescent (red, green, or blue) or biotin tyramide in 1× amplification diluent on each slide. Incubate the slides for approximately 10-30min at room temperature without a coverslip.
- Wash the slides 3 × 5min with TNT at room temperature.
- For detection of biotin tyramide, incubate the slides with steptavidin FITC diluted 1:1000 in blocking solution as described in step 3.
- Wash the slides as described in step 4.
- Dehydrate the slides in ethanol, air dry, and mount in Vectashield.
IV. COMMENTS
The protocol just described allows sensitive detection of specific mRNAs in a variety of cell types, including cultured cells, trypanosomes (Chaves et al., 1998), malaria parasites (Vervenne et al., 1994), and blood cells. The various steps in this protocol have been optimized for detecting RNA molecules in cultured mammalian cells by trial and error (Dirks et al., 1993) and may need some adjustments for specific applications. For example, if RNA FISH is applied at the electron microscopic level of resolution, sample preparation, fixation, and pretreatment conditions
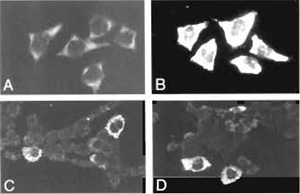
Compared to conventional immunocytochemical detection systems, the use of tyramide-based detection systems results in an at least a 10-fold increase in signal intensity (Raap et al., 1995). This is illustrated in Fig. 1, showing hybridization signals of human elongation factor (HEF) mRNA in HeLa cells and of IL-6 and G-CSF mRNA in human bladder carcinoma cells grown on microscope slides. If required, the localization properties of tyramides can be improved by the addition of dextran sulfate to the amplification diluent (Van Gijlswijk et al., 1996).
This hybridization protocol also allows bright-field microscopic visualization of hybridization signals when conjugates with peroxidase or alkaline phosphatase are used instead of fluorochrome antibody conjugates (Dirks et al., 1993).
As a positive control on the procedure, probes specific for housekeeping gene transcripts, like human elengation factor and actin mRNA, or for rRNA, such as 28S rRNA, can be used.
Finally, in order to study dynamic aspects of RNA localization, methods have been developed that allow hybridization of probes to RNAs and its subsequent visualization in living cells (Molenaar et al., 2001; Pederson, 2001).
V. PITFALLS
- Probe labeling can be checked by performing a filter spot test. Spot 1µl of a dilution series of the labeled probe on a nitrocellulose filter and incubate with sheep antidigoxigenin-alkaline phosphatase. After performing the NBT/BCIP reaction, a probe concentration of 5 to 1 pg/µl should be visible.
- We noticed that it is important to use for the fixation of cells a good source of formaldehyde. Formaldehyde solutions of poor quality may lead to poor cell morphology, high levels of autofluorescence, or weak hybridization signals.
- It is important to titrate the pepsin concentration and/or time of incubation in order to find a balance between morphology and signal intensity.
- For optimal results it is sometimes necessary to denature the target mRNA. This is done just before hybridization, after applying the probe solution and coverslip on the slide, by placing the slide on an 80°C hot plate for 2 to 3 min.
- The dilution and reaction time of labeled tyramides need to be optimized for each application. To diminish nonspecific background staining, the first antibody may be diluted further.
References
Adams, J. C. (1992). Biotin amplification of biotin and horseradish peroxidase signals in histochemical stains.
J. Histochem. Cytochem. 40, 1457-1463.
Chaves, I., Zomerdijk, J., Dirks-Mulder, A., Dirks, R. W., Raap, A. K., and Borst, P. (1998). Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95, 12328-12333.
Dirks, R. W. (1998). Combination DNA/RNA FISH and immunophenotyping. In "Current Protocols in Cytometry" (J. P. Robinson, Z. Darzynkiewics, P. N. Dean, A. Orfao, P. S. Rabinovitch, H. J. Tanke, and L. L. Wheeles, eds.), pp. 8.7.1-8.7.14. Wiley, New York.
Dirks, R. W., Van de Rijke, F. M., Fujishita, S., Van der Ploeg, M., and Raap, A. K. (1993). Methodologies for specific intron and exon RNA localization in cultured cells by haptenized and fluorochromized probes. J. Cell Sci. 104, 1187-1197.
Dirks, R. W., Van Gijlswijk, R. P. M., Vooijs, M. A., Smit, A. B., Bogerd, J., Van Minnen, J, Raap, A. K., and Van der Ploeg, M. (1991). 3'- End fluorochromized and haptenized oligonucleotides as in situ hybridization probes for multiple, simultaneous RNA detection. Exp. Cell Res. 194, 310-315.
Femino, A. M., Fay, F. S., Fogarty, K., and Singer, R. H. (1998). Visualization of single RNA transcripts in situ. Science 280, 585-590.
Levsky, J. F., Shenoy, S. M., Pezo, R. C., and Singer, R. H. (2002). Single-cell gene expression profiling. Science 297, 836-840.
Macville, M. V., Van Dorp, A. G., Dirks, R. W., Fransen, J. A., and Raap, A. K. (1996). Evaluation of pepsin treatment of electron microscopic RNA in situ hybridization on ultra-thin cryosections of cultured cells. Histochem. Cell Biol. 105, 139-145.
Macville, M. V., Wiesmeijer, K. C., Dirks, R. W., Fransen, J. A., and Raap, A. K. (1995). Saponin pre-treatment in pre-embedding electron microscopic in situ hybridization for detection of specific RNA sequences in cultured cells: A methodological study. J. Histochem. Cytochem. 43, 1005-1018.
Molenaar, C., Marras, S. A., Slats, J. C. M., Truffert, J.-C., Lemaitre, M., Raap, A. K., Dirks, R. W., and Tanke, H. J. (2001). Linear 2' O-methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 29, e89.
Pederson, T. (2001). Fluorescent RNA cytochemistry: Tracking gene transcripts in living cells. Nucleic Acids Res. 29, 1013-1016.
Raap, A. K., Van de Corput, M. P. C., Vervenne, R. A. W., Van Gijlswijk, R. P. M., Tanke, H. J., and Wiegant, J. (1995). Ultrasensitive FISH using peroxidase-mediated deposition of biotinor fluorochrome tyramides. Hum. Mol. Genet. 4, 529-534.
Snaar, S. P., Vincent, M., and Dirks R. W. (1999). RNA polymerase II localizes at sites of human cytomegalovirus immediate-early RNA synthesis and processing. J. Histochem. Cytochem. 47, 245-254.
Van de Corput, M. P. C., Dirks, R. W., Van Gijlswijk, R. P. M., Van Binnendijk, E., Hattinger, C. M., De Paus, R. A., Landegent, J. E., and Raap, A. K. (1998). Sensitive mRNA detection by fluorescence in situ hybridization using horseradish peroxidase-labeled oligodeoxynucleotides and tyramide signal amplification. J. Histochem. Cytochem. 46, 1249-1259.
Van Gijlswijk, R. P. M., Wiegant, J., Raap, A. K., and Tanke, H. J. (1996). Improved localization of fluorescent tyramides for fluorescence in situ hybridization using dextran sulphate and polyvinyl alcohol. J. Histochem. Cytochem. 44, 389-392.
Vervenne, R. A. W., Dirks, R. W., Ramesar, J., Waters, A. P., and Janse, C. J. (1994). Differential expression in blood stages of the gene coding for the 21-kilodalton surface protein of ookinetes of Plasmodium berghei as detected by RNA in situ hybridization. Mol. Biochem. Parasitol. 68, 259-266.