Enzymology and Evolution of the SMT
SMTs are a family of AdoMet-dependent C-methyltransferases that use two substrates, AdoMet as a methyl source and sterols with a Δ24-bond as the acceptor molecules, for the transmethylating reaction yielding AdoHcy and phytosterols with single or double methylation at C-24. AdoMet-dependent methylations are important in generating phytosterols as primary metabolites, and other AdoMet-dependent methylations contribute to generating many secondary products, including phenylpropanoids, flavonoids, and alkaloids (Nes et al., 1986; Ounaroon et al., 2002; Schubert et al., 2003; Zubieta et al., 2002). The primary structures and enzyme kinetics as well as the requirements for the substrate may be quite different among the methyltransferases. However, a related evolution may be inferred to the generation of these seemingly different plant enzymes in which the active center evolved a common core of structurally similar amino acid residues for interaction with AdoMet.SMT-catalyzed reactions are remarkable in that they convert lipophilic compounds to methylated olefins in a single step. The details of these processes have intrigued scientists for half a century. A comparison of
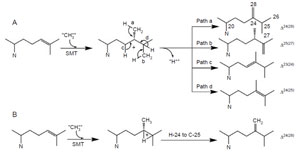 |
| FIGURE 9.13 Possible C-methylation mechanisms for producing C-28 olefins. (A) Stepwise or carbocationic pathway; (B) nonstop or concerted pathway. |
biomimetic studies of uncatalyzed versus SMT-catalyzed reactions reveal that the elementary chemical steps required for C-methylation of an olefin can take place to produce multiple products in a nonezymatic reaction following predictable chemical principles (Julia and Marazano, 1985; Venkatramesh et al., 1996). Mechanistically, the key to the C-methylation reaction is the positively charged sulfoniumcenter in AdoMet which renders the methyl group electrophilic and susceptible to the relatively nucleophilic Δ24-double bond. The ensuing reaction proceeds by a reorganization of at least three bonds: (1) cleavage of the C–S bond in the AdoMet donor, (2) formation of the C-24(28)-bond between the donor and the acceptor, and (3) loss of a proton from either the donor or acceptor. SMT catalysis proceeds stereo- and regiospecifically to generate distinct product sets by variations of a similar ionic mechanism (Fig. 9.13). Together these enzymes are capable of converting sterol precursor into more than 100 distinct phytosterols in plants (Nes and McKean, 1977).
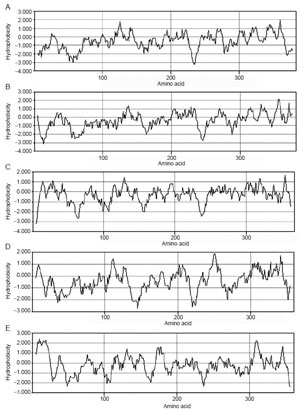 |
| FIGURE 9.14 Hydropathy plots corresponding to SMTs for (A) S. cerevisiae; (B) Gibberella fujikuroi; (C) Glycine max; (D) Trypanosoma brucei; (E) Arabidopsis thaliana. |
SMTs are membrane-bound enzymes which share a high degree of similarity in primary structure as revealed by a comparison of their amino sequences reported in the GenBank. Hydropathy analyses of SMTs from different organisms indicate that they are similar and moderately hydrophobic with no membrane spanning domains (Fig. 9.14). Several cloned SMT enzymes from plants, fungi, and protozoa have been overexpressed in Escherichia coli (Nes et al., 1998b, 2003; Zhou et al., 2006). The open reading frame of these catalysts will code for predicted proteins of 336–383 amino acids with a molecular mass that ranges from 38.5 to 43.3 kDa. In all cases studied, the purified protein possesses a tetrameric subunit organization that ranges from 160 to 172 kDa. Equilibrium dialysis and Scatchard plotting of Kd measurements indicate that the SMT has a single binding site for sterol and AdoMet. Catalytic constants for the native substrates for these enzymes are generally Km ca. 30 µM and kcat ca. 0.01 s —1. The secondary structure of several SMT1 enzymes, as determined by circular dichroism, was found to be similar (Nes et al., 2004; Zhou and Nes, 2003). In the case of the yeast SMT, the following population of structures was recorded: 43% α-helix, 29% β-sheet, 7% turn, and 21% random coil (Zhou and Nes, 2003). These findings suggest the conformational features of the native SMTs will be similar. Although X-ray crystallographic analysis of heavy atom-labeled crystalline SMT bound to a suicide substrate could provide much helpful information about structure–function relationships, crystallization of the SMT has proven to be a formidable task and no threedimensional structure of the enzyme is available to date. Despite this fact, sufficient activity assays of substrate and transition state analogs with different SMTs are available to speculate a steric–electric model of SMT catalysis (Fig. 9.15) (Parker and Nes, 1992).
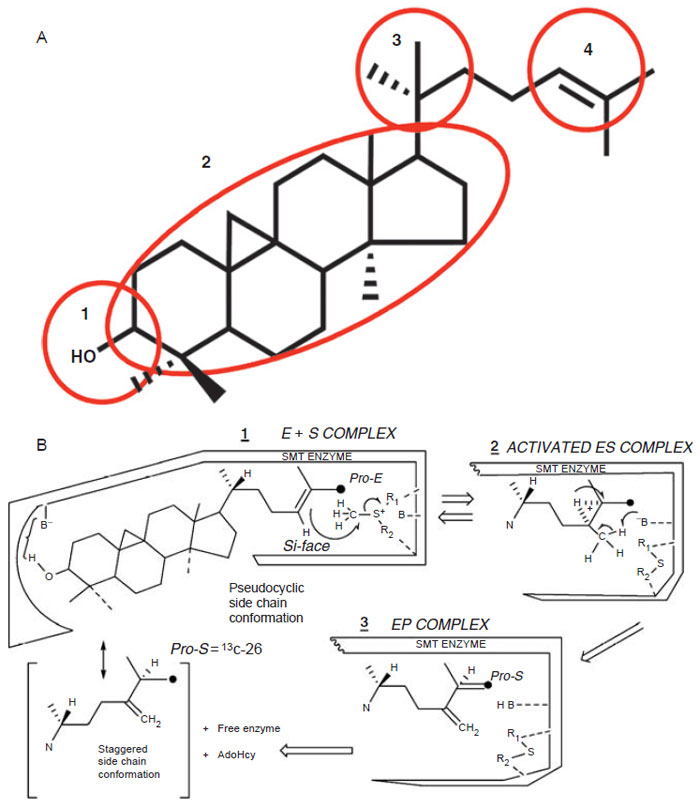 |
| FIGURE 9.15 Postulated domains of sterol molecule (Top, A) recognized by the SMT and the steric–electric plug model of SMT catalysis (Bottom, B). (See Page 12 in Color Section.) |
The model predicts that four domains of the sterol molecule are critical to productive binding and catalysis. Domain 1 recognizes an equatorial C3–OH group that is nucleophilic to bind to a polar amino acid; Domain 2 recognizes a double bond in the nucleus, preferable at Δ8(9) (or a 9β,19 cyclopropane group), to lock the nucleus in a pseudoplanar conformation thereby generating a flat shape and interacts with the angular methyl groups at C-10 and C-13 to secure the sterol in the hydrophobic cleft; Domain 3 recognizes a 20R-configuration to direct the side-chain into a ‘‘right-handed’’ conformation thereby positioning C-22–C-26 to a catalytically acceptable conformation; and Domain 4 recognizes a side-chain that contains the terminal C-25-C-26–C-27-isopropyl group and Δ24-bond to anchor the side-chain near AdoMet. The model also predicts a conformational change in the enzyme during catalysis to allow for the different kinetics of the reaction (Parker and Nes, 1992).
For the first C1-activity catalyzed by the yeast SMT, the methyl transfer reaction proceeds by a simple SN2 reaction involving a dative bond formed between C-24 and C-28 and a bridged carbenium ion formed opposite to the side of the original double bond facing the AdoMet. A 1,2-hydride shift of H-24 to C-25 proceeds because both C-24 and C-25 have become tertiary sites, equally stabilized by hyperconjugation. The hydride shift of H-24 to C-25 and methyl transfer (to C-24 from AdoMet) are concerted; only in this way will there be no formation of a discrete C-24 or C-25 cation and channeling will be restricted to formation of a single product (Nes et al., 1998b; Nes et al., 2003).
The plant SMT1, in contrast to the yeast SMT1, can catalyze both the first and second C1-transfer reaction. The first C1-activity acts mechanistically similar to the yeast SMT1 to produce a Δ24(28)-product. However, the second C1-activity can generate a mixture of 24-ethyl sterols by an ionic mechanism involving a C-24 isofucosterol cation and a reversible 1,2-hydride shift of H-24 to C-25 that leads to deprotonation and formation of different olefins corresponding to the side-chains of fucosterol (Δ24(28)E), isofucosterol (Δ24(28)Z), and clerosterol (Δ25(27) 24β-ethyl) (Nes et al., 2003). The chemical mechanisms associated with the first and second C1-activities differ in the order the two bonds are cleaved in the Δ 24(25)- and Δ24(28)-substrates. In the carbocation mechanism, the bond to the leaving group is broken first whereas in the concerted mechanism, both bonds are cleaved simultaneously without intervention of an intermediate. Thus, the second C1-activity can operate a stepwise mechanism whereas the first C1-activity can operate a nonstop mechanism. In similar fashion, the first C1-transfer can proceed by a stepwise mechanism to produce multiple products, as recently found for the cloned Trypanosoma brucei SMT that converts the Δ24(25)- sterol to a mixture of Δ24(28)-, Δ25(27)-, and Δ24(25)-sterols (Zhou et al., 2006). The operation of path a in the yeast SMT compared to the operation of paths a and b versus path d in the T. brucei SMT (Fig. 9.13) suggests that the yeast SMT can operate a 1-base mechanism whereas the T. brucei SMT can operate a 2-base mechanism for the coupled methylation–deprotonation reaction.
Studies on a set of cloned wild-type and mutant yeast SMT (Nes et al., 1999; Nes et al., 2002; Zhou and Nes, 2003) indicate that the conserved acidic amino acids at D125 and D152 form a wall of the AdoMet binding site, perhaps hydrogen bonding to the methionine and ribose moieties of the substrate; D276 and E195 are positioned directly or by way of a water bridge to the proximal (C3-hydroxyl group of the sterol) and distal (Δ24-bond of the sterol) nucleophilic segments of the acceptor, respectively. E195 may also interact with the positive charge on the sulfur residue of AdoMet in which case it may serve as a counterion to AdoMet. Alternatively, through cation-π interactions, a neighboring aromatic amino acid may serve as the counterion to AdoMet. H90, positioned above (Si-face of the 24,25-double bond) the substrate double bond in the same plane as AdoMet, may serve as the base involved with C-28 deprotonation that leads CH3 to CH2 production. The residue at Y81 may lie on the Re-face of the substrate double bond and act during the methylation-deprotonation reaction to stabilize the high-energy intermediate(s) formed during the reaction progress. Negatively charged ions included in the SMTs are considered to be arranged in the active site so as to stabilize the intermediary carbocations formed during catalysis through cation-π interactions (Nes et al., 2004), thereby restricting channeling and leading to an acceleration of the C-methylation reaction. Homology modeling and the enzymatic studies with SMT predict a spatial arrangement of the secondary structural elements in relation to sterol and AdoMet substrates as shown in Fig. 9.16.
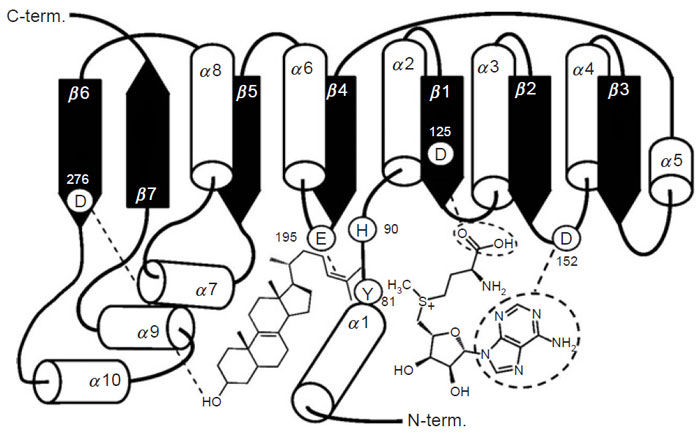 |
| FIGURE 9.16 Schematic representation of the methyltransferase fold of the SMT; spatial arrangement of the secondary structure elements in relation to sterol and AdoMet substrates. Adapted from Nes et al., 2004. |
Recent developments in the cloning and purification of SMTs and studies performed in our laboratory (Mangla and Nes, 2000; Nes, 2000; Nes et al., 2003; Zhou et al., 2006) and that of Benventiste (2004) have led the enzyme commission (EC) to reclassify the SMT family according to substrate preference into three classes: the fungal SMT1 prefers zymosterol [SMT1ZY], EC 2.1.1.41; the plant SMT1 prefers cycloartenol [SMT1CA], EC 2.1.1.142; and the plant SMT2 prefers 24(28)-methylene lophenol [SMT2ML], EC 2.1.1.143. SMT1 and SMT2 catalyze the first and second methylation activities, respectively. If two enzymes belong to the same class in this classification, they are considered to have similar chemical functions. To date, the EC does not recognize the ability of SMTs to catalyze different product distributions as a measure of function. Moreover, there must be additional SMTs with substrate preferences not heretofore recognized by the EC. For example, fungi exist that accumulate lanosterol rather than zymosterol when treated with an inhibitor of SMT (Nes et al., 2002), suggesting these catalysts, referred to as SMT1LA, prefer lanosterol to zymosterol.
The steric–electric plug recognizes structural complementarity between the SMT and the substrate molecules. However, the substrate affinity and enzymatic product are not always either obvious or predictable for an unknown SMT. For example, the plant SMT1CA from algae and the protozoan SMT1ZY can generate similar 24-methyl-Δ25(27)-olefins as the major product. Alternatively, the fungal SMT1ZY from yeast and the plant SMT1CA from soybean can generate similar 24 (28)-methyl(ene)-Δ24(28)-olefins. The first C1-activity of plant SMT2 catalyzes cycloartenol to a single methylated product whereas the second C1-activity catalyzes 24(28)-methylene lophenol to three methylated products. The ability of the plant SMT2 to catalyze substrates of different features to different product sets was shown to result from the specificity in molecular recognition of the Δ24-sterol structure. Kinetically, the SMT1 from fungi and plants differ. In the case of the yeast SMT1, the mechanism is random and for the plant SMT1 ordered so that AdoMet is the leading substrate and must bind before sterol to the enzyme (Nes, 2000; Nes et al., 2003). Most likely, the different kinetic mechanisms relate to the significance that the plant SMT1 has activities for both the first and second C1-transfer reaction and an ordered mechanism enables sequential C1-transfer of different acceptors in the active site.
SMT1 and SMT2 activities vary with respect to one another in plants (Nes, 2000; Nes et al., 1989b; Wentzinger et al., 2002). For example, the proportion of SMT1 to SMT2 activity measured as their catalytic competence (Vmax/Km) is similar in cultured cells of tobacco during active cell proliferation. Alternatively, as the plant matures and cell proliferation is arrested, the ratio of the two activities change dramatically and can favor the activity of the second C1-transfer reaction. These findings are in agreement with the suggestion of Benveniste and coworkers (Arnqvist et al., 2003; Benveniste, 2004) of the possible importance of SMT2 as a branch point enzyme in phytosterol synthesis. The activity of SMT1 to control carbon flux into the phytosterol pathway and to cholesterol (Diener et al., 2000; Parker and Nes, 1992) is evidenced in the high specific activity of cholesterol synthesis in mature leaves (Nes, 1990) and in potato plants overexpressing a foreign SMT1 (Fonteneau et al., 1977). Given the regulatory role of SMT1 and SMT2 to balance the ratio of C-8 to C-9 to C-10 sterols suggests that the induced change in the ratio of SMT1 to SMT2 activities during development is largely controlled at the level of transcription. However, effectors, such as sitosterol or ATP, may also increase or decrease carbon flux through a branch point in phytosterol synthesis by modulating SMT activity (Nes, 2000).
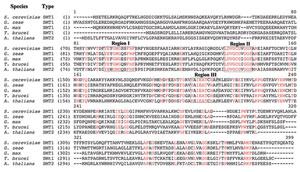
AdoMet-dependent methylation has been found to be the target of functional convergence (Schubert et al., 2003) generating a subfamily of SMTs that clearly evolved their substrate specificity for sterol independently of related AdoMetdependent methyltransferases. Genetic and bioinformatic research of the derived amino acid sequences of over 60 SMTs deposited in the GenBank and other databases, indicate the primary structure encodes about 380 (±20) amino acid long proteins with an amino acid sequence relatedness which allows subdivision of the SMT gene family into five subfamilies, designated SMTa through SMTe, each distinguished by sharing a minimum of 40% identity among members (Fig. 9.17). Certain sequence motifs shared by all SMTs suggest that they share a common evolutionary origin. For example, three conserved domains in the primary structure of SMTs, which we have previously designated as Regions I, II, and III, are hydrophobic and always found in the same order on the polypeptide chain and are separated by comparable intervals (Fig. 9.18). Chemical affinity labeling and site-directed mutagenesis experiments have shown that Regions I and III correspond to a sterol binding site and in the yeast SMT maps to Y81EYGWGSSFHF and Y192AIEATCHAP (Nes et al., 1999; Sinha, 2004; Zhou and Nes, 2003). Photoaffinity labeling and site-directed mutagenesis experiments show that Region II corresponds to a AdoMet binding site and in the yeast SMT maps to L24DVGCGVGGP (Schaeffer et al., 2000). That these enzymes are so similar in much of their primary and presumably three-dimensional structures in spite of taxonomic differences in the species from which they are derived suggests that they arose by divergent evolution from an ancestral gene prior to the origin of eukaryotes.
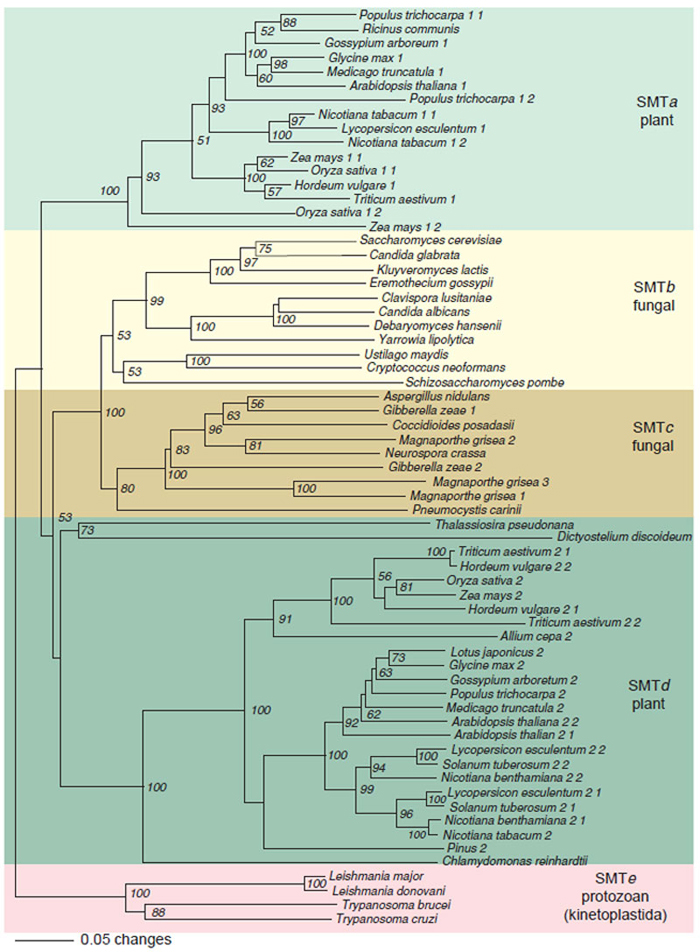 |
| FIGURE 9.17 Rooted phylogenetic tree of eukaryotic SMT was created with PAUP using the neighbor-joining method with kinetoplastida SMTs as the out group. The scale bar represents a distance of 0.05 substitutions per site. Numbers are the percentage bootstrap values for 1,000 replicates. SMTa through SMTe designate SMT subfamilies defined by a minimum of 35% identity between members at the amino acid level. Accession number of SMT sequences obtained from NCBI (http://www.ncbi.nlm.nih.gov): Ricinus communis, T10173; Glycine max 1, T06780; Arabidopsisthaliana 1, AGG28462; Nicotiana tabacum 1-1, AAC43951; Nicotiana tabacum 1-2, AAC35787; Zea mays 1-1, TO4138; Oryza sativa 1-1, AAC34988; Triticum aestivum 1, ABB49388; Oryza sativa 1-2, AAP21419; Saccharmyces cerevisiae, NP_013706; Candida glabrata, CAG59930; Kluyveromyces lactis, AAS52116; Clavispora lusitaniae, CAO21936; Candida albicans, O74198; Debaryomyces hansenii, CAG87427; Yarrowia lipolytica, CAG77980; Ustilago maydis, EAK84412; Schizosaccharomyces pombe, CAB16897; Gibberella zeae 1, XP_382959; Magnaporthe grisa 2, EAA48309; Neurospora crassa, CAB97289; Gibberella zeae 2, XP_355916; Magnaporthe grisa 3, EAA50587; Magnaporthe grisa 1, EAA47049; Pneumocystis carinii, AKK54439; Oryza sativa 2, ACC34989; Arabidopsis thaliana 2-1, ABB62809; Arabidopsis thaliana 2-2, CAA61966; Nicotiana tabacum, TO3848; Leishmania donovani, AAR92098; Trypanosoma cruzi, TIGR_5693. Gene indices of SMT obtained from TIGR (http://www.tigr.org): Populus trichocarpa 1-1, TC35619; Medicago trucatula, TC86500; Gassypium arboretum 1, TC20798; Populus trichocarpa 1-1, TC36329; Hordeum vulgare 1, TC 110279; Zea mays 1-2, TC234797; Cryptococcus neoformans, TC4573; Aspergillus nidulans, TC6295; Coccidioides posadasii, TC4513; Triticum aestivum 2-1, TC165856; Hordeum vulgare 2–2, TC121611; Zea mays, TC224796; Hordeum vulgare 2-1, TC123636; Triticum aestivum 2-2, TC172448; Allium cape 2, TC2207; Lotus japonicus, TC7995; Glycine max 2, TC189052; Gossypium arboretum 2, TC 21049; Populus trichocarpa 2, TC36329; Medicago truncatula 2, TC77751; Lycopersicon esculentum 2-2, TC126730; Solanum tuberosum 2-2, TC126449; Nicotiana benthamiana 2-2, TC7251; Lycopersicon esculentum 2-1, TC124648; Solanum tuberosum 2-1, TC112127; Nicotiana benthamiana 2-1, TC8230; Pinus 2, TC52326; Chlamydomonas reinhardtii, TC29837. Gene identification number of SMT from The Wellcome Trust Sanger Institute (http://www.sanger.ac.uk): Leishmanis major, LM3731Bb05.p1c; Trypanosoma brucei, TB10.1520. The SMT gene of Dictyostelium discoideum was from IMB (http://genome.imb-jena.de) and gene identification number is pcr25kl1p3887. SMT gene of Thalassiosira pseudonana is identified from GRI (http://genome.jgi-psf.org) in scaffold_30, 94659:96009. (See Page 13 in Color Section.) |
To understand further the importance of conserved amino acids in the primary structure and the molecular interactions between sterol and enzyme to correctly position and discriminate the nucleophile that serves to undergo C-methylation to a single product, mutagenesis and activity assay with carefully designed mechanismbased inactivators were undertaken of highly conserved amino acid residues in the SMT from yeast (Fig. 9.19). We have reported that several mutants of yeast SMT modified in Region I, D79L and Y81F, produce mixtures of Δ24(28)- and Δ25(27)- olefins depending on the nature of Δ24-substrate (Nes et al., 1999; Sinha, 2004; Zhou and Nes, 2003). The yeast SMT1 can become plant-like in accepting Δ24(28)-substrates by a single mutation in Region I at Y81 (Nes et al., 1999). The suicide substrate, 26,27-dehydrozymosterol, assayed with wild-type enzyme produces a novel Δ23-sterol with an elongated side-chain (Parker and Nes, 1992). The steric–electric plug model permits differences in product specificity among SMT enzymes to have arisen through point mutations which change either the shape of the catalytic site and/or positions of the
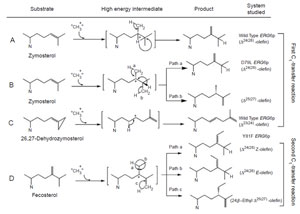 |
| FIGURE 9.19 Catalytic competence of native and mutant yeast SMTs tested with different substrates. |
crucial functional groups. The D79L and Y81F mutants are capable of overcoming the topological impediment to generate multiple products, suggesting similar binding segments in the active center are present in all SMTs.
Testing with different sterol substrates of either heterologously expressed or wild-type enzymes indicates that substrate specificities have evolved differently in plants compared to the fungi and protozoa. All plant SMT1s reveal a strict substrate specificity accepting cycloartenol whereas the fungal SMT1 and protozoan SMT1 accept either zymosterol or lanosterol. A major difference between zymosterol and lanosterol is the presence of the geminal methyl group at C-4 in the lanosterol structure. The 4,4-dimethyl group can sterically interfere with the hydrogen bonding ability of the C3-hydroxyl group thereby affecting substrate affinity. Whereas SMT1 enzymes can be distinguished on their recognition of the nucleus structure, SMT1 and SMT2 enzymes can be distinguished on their recognition of the side-chain functional group Δ24(25)- versus Δ24(28)-substrate. To account for the different substrate acceptability, different arrangements of similar amino acid residues may have evolved in the active site of SMTs to interact with the nucleophilic groups at C-3 and C-24.
No conclusive discussion for the order of SMT evolution could be drawn from the unrooted phyogenetic tree (Fig. 9.17), based on the order of intermediates and the positioning of SMT isoforms in phytosterol synthesis. However, plant SMT2s which utilize 24(28)-methylene lophenol as the preferred substrate, but maintain vestige substrate specificity for cycloartenol, must have evolved from a progenitor plant SMT1 perhaps by duplication followed by mutation and divergence. An important question that warrants further study is whether cycloartenol served as a template providing functional constraints in the design and genetic formation of the plant SMT active site during either evolution or random mutation with natural selection provided the guiding principle of enzyme redesign.
A similarity in function (catalytic competence) among SMTs is reflected in the enzyme ability to catalyze a sterol acceptor to a methylated product. However, enzyme function of individual SMT isoforms in either the same plant or those formed in different phyla can be different in terms of substrate acceptability, kinetics, product outcome, and selectivity to inhibitors or effectors of SMT action. Although it may be possible to use the conserved Regions I and II of cDNA corresponding to SMT for homology-based cloning strategies, there is insufficient information for predictions of catalysis of new SMTs that might be cloned from, for example prokaryotes, since none of the putative prokaryote SMTs in the GenBank contain Region I. The trace level of phytosterol in cyanobacteria, suggesting they are contaminants (Marshall, 2007), and the likelihood SMTs are not synthesized by photosynthetic bacteria make the timing of SMT production and hence phytosterol accumulation during the course of evolution unclear.