In view of this if the length of DNA is known, then the maximum length of protein which can be synthesized, can be worked out in terms of number of amino acid residues. For instance, a DNA segment of 1200 base pairs can give rise to a protein having not more than 400 amino acids. Moreover, if number of amino acids in proteins synthesized, and number of nucleotide base pairs in DNA are known, they can be compared and one would expect that the number of base pairs should be at least 3 times the number of amino acids in the protein. Rather if there are segments of DNA which are not coding for protein, then the length of DNA segment will be more than three times the number of amino acid residues in protein. Such comparisons have been made in bacteriophage ΦX174 which contains a single stranded DNA approximately 5,400 nucleotides in length. Complete sequence of these 5,400 nucleotides was also worked out by
Fredrick Sanger and his colleagues at the MRC Laboratory of Microbiology, Cambridge, U.K. (Fredrick Sanger for this and other work got the second Nobel Prize in the year 1980).
The genome of ΦX 174 consists of nine cistrons. Amino acid sequences of major proteins coded by the genome of ΦXl74 have also been worked out and their molecular weights arc known. From the information about proteins coded, an estimate could be made of the number of nucleotides required. This estimate of number of nucleotides exceeds 6,000 which is much higher than the actual number of nucleotides present i.e., 5,400. Therefore, it was difficult to explain how these proteins could by synthesized from a DNA segment which is not long enough to code for the required number of amino acids. On detailed study of the system, it was discovered that sequences in the same segment could be utilized by two different cistrons coding for different proteins. Such overlapping of cistrons will be theoretically possible if the two cistrons have to function at different times and their nucleotide sequences are translated in two different reading frames.
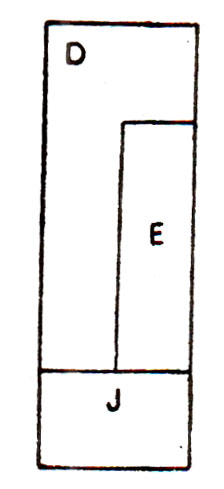
Fig. 29.13. Overlapping of E and D genes in phage ΦX174.
In 1976
Barrell and his co-workers discovered that in ΦXl74, having nine cistrons (A, B, C, D, E, J, F, G, H), cistron E is present between D and J and that the cistron E overlaps cistron D. This could be done by working out the nucleotide sequences of DNA segment representing gene D, E, and J. It could be shown that amber mutations in cistron E lie within the cistron D and these amber mutations do not influence the translation of cistron D into its protein. Similarly some other nonsense mutations for cistron E also lie in cistron D suggesting that the cistrons D and E overlap in the DNA sequences and that the cistron D and E are translated in two different reading frames so that amber codon in mRNA of one cistron will not be read as termination codon during the translation of mRNA of the other cistron.
Among genes D, E, and J in ΦXl74, cistron D codes for a simple protein, cistron E codes for a protein for lysis and cistron J codes for a particle component. It was shown that enough space is not available where these three genes are located, thus confirming that there was overlapping between D and E cistrons. Overlapping has also been suggested to be present in regions of genes A and B and mutants were located in overlapping zone. Overlapping regions were also found to be present at beginnings of cistrons A, C, and J, where in each case the initiation codon of a cistron overlaps with the termination codon of its corresponding previous cistron. A DNA segment showing overlapping of D and E is shown in Figure 29.13.

Fig. 29.13. Overlapping of E and D genes in phage ΦX174.
Overlapping sequences have also been discovered in tryptophan mRNA of
E. coli and several other cases. In view of wide occurrence of overlapping genes, it seems that phenomenon of overlapping genes, is an economic device to make better use of genetic material, through packing of more genetic information in less DNA. This phenomenon may be of general occurrence. Therefore, it may be necessary to modify our concept of coding potential (number of amino acids, which a DNA segment can code for) of DNA genomes, because a DNA can potentially code for many more amino acids in different reading frames than the number of amino acids expected to be coded, if no overlapping was present.





