Techniques used in recombinant DNA
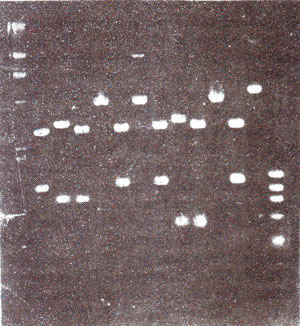
Separation of DNA fragments using agarose or poiyacrylamide gel electrophoresis. When genomie DNA, extracted from any tissue of a plant or animal species, is digested with a restriction enzyme, it is cleaved into segments. The, segments of different sizes can be separated through gel electrophoresis before a molecular probe is used to detect the segments which have sequences similar to those in the probe (for molecular probes, see later in this section). Gel electrophoresis involves movement of fragments or molecules under a well created on one edge of the gel. The gel may be a cylinder or a slab (usually a slab for cloning expt.), about 10 cm long and 0.5 cm thick.
The rate of movement of fragments is inversely correlated with the size of fragments or molecules, so that heavier fragments will remain closer to the site of loading and the lighter fragments will move away (Fig. 39.1). Fragments of different sizes will appear as bands on the gel and can be examined or isolated for further study. More often agarose gels are used, but for separation of fragments differing by few base pairs, polyacrylamide gels are used. Polyacrylamide gels are more commonly used for DNA sequencing experiments (see Genetic Engineering and Biotechnology 3. Isolation, Sequencing and Synthesis of Genes).
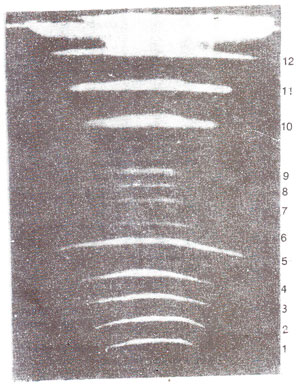 Separation of large DNA molecules, (whole chromosomes) using PFGE. The technique of gel electrophoresis is used for separation of DNA molecules of different sizes. However, DNA molecules of large size could not be handled in this techniques. In recent years, a new technique called pulsed field gel electrophoresis (PFGE) has been used, for separation of large sized DNA molecules, sometimes representing whole chromosomes. Using this technique, separation of DNA molecules belonging to each of the 16 individual chromosomes of yeast has become possible (Fig. 39.2). In this technique, short pulses of electricity are used in two different directions and DNA is embedded and used in the form of agarose plugs to avoid fragmentation of large DNA molecules. Using the technique of PFGE and a more refined technique CHEFE (contour clamped homogeneous electric field electrophoresis) genomes of several fungi could be resolved into chromosomal bands and used for mapping of DNA sequences on specific chromosomes.
Separation of large DNA molecules, (whole chromosomes) using PFGE. The technique of gel electrophoresis is used for separation of DNA molecules of different sizes. However, DNA molecules of large size could not be handled in this techniques. In recent years, a new technique called pulsed field gel electrophoresis (PFGE) has been used, for separation of large sized DNA molecules, sometimes representing whole chromosomes. Using this technique, separation of DNA molecules belonging to each of the 16 individual chromosomes of yeast has become possible (Fig. 39.2). In this technique, short pulses of electricity are used in two different directions and DNA is embedded and used in the form of agarose plugs to avoid fragmentation of large DNA molecules. Using the technique of PFGE and a more refined technique CHEFE (contour clamped homogeneous electric field electrophoresis) genomes of several fungi could be resolved into chromosomal bands and used for mapping of DNA sequences on specific chromosomes.
Southern, Northern and Western blotting. A mixture of DNA, or RNA or protein fragments can be separated by gel electrophoresis and the separated bands can be stained and visualized directly in the gel. However, to confirm the identity of these bands or to find similarity of one or more of these bands with a known and available molecular probe, it is possible to hybridize these bands with a labelled probe. To facilitate this hybridization, the bands are often transferred to a nitrocellulose membrane through a technique described as blotting. When DNA bands are thus blotted, it is called Southern blotting (after the name of E.M. Southern; when RNA bands are thus transferred it is described by the jargon term Northern blotting and similarly when protein bands are transferred, the technique is described as Western blotting.

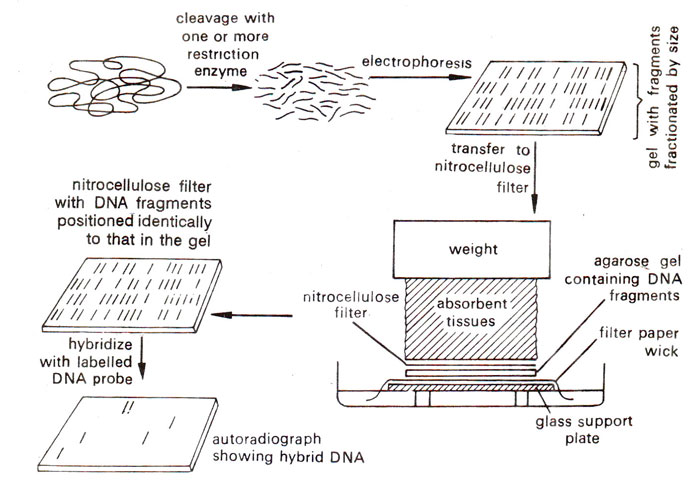 (a) Southern blotting. For Southern blotting DNA sample is first digested with a restriction enzyme and digested sample is gel electrophoresed. The DNA bands in the gel are denatured into single strands with the help of an alkali solution. Subsequently, the gel is laid on top of a buffer saturated filter paper, placed on a solid support (e.g. glass plate), with its two edges immersed in the buffer. A sheet of nitrocellulose membrane is placed on top of the gel and a stack of many papers (paper towels) on top of this membrane. A weight of about 0.5 kg. is placed on top of paper towels. The buffer solution is drawn up by filter paper wick, and passes through the gel to the nitrocellulose membrane and finally to the paper towels.
While passing through the gel, the buffer carries with it single stranded DNA, which binds on to the nitrocellulose membrane, when the buffer passes through it to the paper towels. After leaving this arrangement for a few hours or overnight, paper towels are removed and discarded. The nitrocellulose membrane with single stranded DNA bands blotted on to it, is baked at 80°C for 2-3 hours to fix the DNA permanently on the membrane. This membrane now has a replica of DNA bands from agarose gel, and can be used for hybridization with labelled DNA or RNA probe. The membrane may then be washed to remove any unbound DNA and X-ray film is exposed to the hybridized membrane to get autoradiographs, The above steps involved in Southern blotting are shown in Figure 39.3.
(a) Southern blotting. For Southern blotting DNA sample is first digested with a restriction enzyme and digested sample is gel electrophoresed. The DNA bands in the gel are denatured into single strands with the help of an alkali solution. Subsequently, the gel is laid on top of a buffer saturated filter paper, placed on a solid support (e.g. glass plate), with its two edges immersed in the buffer. A sheet of nitrocellulose membrane is placed on top of the gel and a stack of many papers (paper towels) on top of this membrane. A weight of about 0.5 kg. is placed on top of paper towels. The buffer solution is drawn up by filter paper wick, and passes through the gel to the nitrocellulose membrane and finally to the paper towels.
While passing through the gel, the buffer carries with it single stranded DNA, which binds on to the nitrocellulose membrane, when the buffer passes through it to the paper towels. After leaving this arrangement for a few hours or overnight, paper towels are removed and discarded. The nitrocellulose membrane with single stranded DNA bands blotted on to it, is baked at 80°C for 2-3 hours to fix the DNA permanently on the membrane. This membrane now has a replica of DNA bands from agarose gel, and can be used for hybridization with labelled DNA or RNA probe. The membrane may then be washed to remove any unbound DNA and X-ray film is exposed to the hybridized membrane to get autoradiographs, The above steps involved in Southern blotting are shown in Figure 39.3.

 (b) Northern blotting.Initially the technique of Southern blotting used for DNA transfer from gel to the membrane could not be used for blot-transfer of RNA. Instead mRNA bands from the gel were blot-transferred into a chemically reactive paper, prepared by diazotization of aminobenzyloxymethyl paper. The technique being related to Southern blotting was called Northern blotting (not after name of any scientist as in Southern blotting). Later, it was shown that mRNA bands can be blotted directly onto nitrocellulose membrane, a technique which has been widely adopted. The mRNA bands blotted onto nitrocellulose membrane can be hybridized with a labelled PNA or RNA probe. The single stranded regions of the probe are removed by nuclease (mungbean nuclease or S-l nuclease), so that quantitative estimation of hybridized mRNA can also made.
(b) Northern blotting.Initially the technique of Southern blotting used for DNA transfer from gel to the membrane could not be used for blot-transfer of RNA. Instead mRNA bands from the gel were blot-transferred into a chemically reactive paper, prepared by diazotization of aminobenzyloxymethyl paper. The technique being related to Southern blotting was called Northern blotting (not after name of any scientist as in Southern blotting). Later, it was shown that mRNA bands can be blotted directly onto nitrocellulose membrane, a technique which has been widely adopted. The mRNA bands blotted onto nitrocellulose membrane can be hybridized with a labelled PNA or RNA probe. The single stranded regions of the probe are removed by nuclease (mungbean nuclease or S-l nuclease), so that quantitative estimation of hybridized mRNA can also made.
(c) Western blotting. This technique is used to detect proteins of a particular specificity. Particularly when a transferred gene expresses in transformed cells, the translated product in the form of protein can be identified by this technique. The extracted proteins are subjected to polyacrylamide gel electrophoresis (PGE) and are then transferred onto nitrocellulose, to which they bind. Nitrocellulose membrane is then used for probing with a specific labelled antibody (antibody will not hybridize with protein, but bind with it). The antibody may be labelled with125 I and the signal is detected again with autoradiography. If radioactive label is not used, bound antibody may be detected by a second antibody tagged with an enzyme.
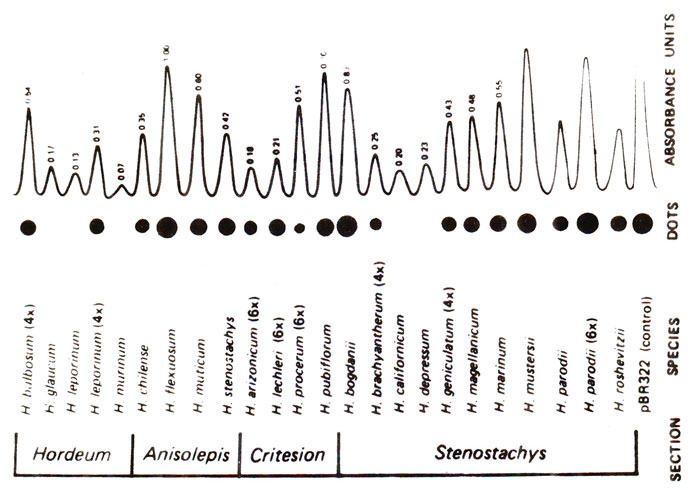
Dot blots and slot blots. Another variety of Southern or Northern blots is dot blots or slot blots. In dot blots, cloned or pure extracted DNAs to be tested are spotted adjacent to each other on a nitrocellulose membrane. DNA blots thus produced are immobilized and denatured so that they are bound on the membrane as single stranded DNA blots. The membrane is then hybridized with radioactively labelled probe (DNA or RNA). The dot representing sequence related to probe will light up by autoradiography (Fig. 39.4). The intensity of the dot will indicate the relative concentration of the probe in the DNA sample used for a particular dot. No steps involving digestion with enzyme, gel electrophoresis or transfer from get to membrane are needed in this technique. Therefore, it is considered much more convenient, when no restriction sites are needed to be studied.
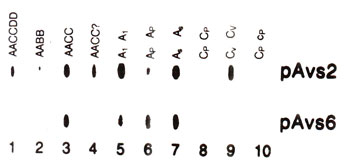
Sometimes, an apparatus is used for placing spots on the membrane, through slots made in this equipment. The spots made thus are in the form of oblong slots rather than round blots. These slots are used just like dot blots and are described as slot blots (Fig. 39.5).

Fig. 39.2. A gel with 12 bands representing IS chromosomes. of yeast as resolved by PFGE (the three doublet bands i.e. 5, 10 and 11 can be resolved on other gels). (From Maynard Olson).

Fig. 39.2. A gel with 12 bands representing IS chromosomes. of yeast as resolved by PFGE (the three doublet bands i.e. 5, 10 and 11 can be resolved on other gels). (From Maynard Olson).

Fig. 39.3. Southern blotting method of analysing DNA segments that share homology with a nucleic acid probe.

Fig. 39.3. Southern blotting method of analysing DNA segments that share homology with a nucleic acid probe.

Fig. 39.4. Dot blots from 25 Hordeum species, showing difference in abundance of sequence related to the probe pSC119.

Fig. 39.5 Slot blots from genomic DNA of 10 Avena species, showing difference in abundance of sequences related to two different probes (pAvs2, pAvs 6).
Dot blots and slot blots. Another variety of Southern or Northern blots is dot blots or slot blots. In dot blots, cloned or pure extracted DNAs to be tested are spotted adjacent to each other on a nitrocellulose membrane. DNA blots thus produced are immobilized and denatured so that they are bound on the membrane as single stranded DNA blots. The membrane is then hybridized with radioactively labelled probe (DNA or RNA). The dot representing sequence related to probe will light up by autoradiography (Fig. 39.4). The intensity of the dot will indicate the relative concentration of the probe in the DNA sample used for a particular dot. No steps involving digestion with enzyme, gel electrophoresis or transfer from get to membrane are needed in this technique. Therefore, it is considered much more convenient, when no restriction sites are needed to be studied.