cDNA library from mRNA
Complementary DNA (cDNA) libraries can also be prepared by isolating mRNA from tissues which are actively synthesizing proteins, like roots and leaves in plants, ovaries or reticulocytes in mammals, etc. The mRNA is used for copying it into cDNA through the use of reverse transcriptase.
A cDNA molecule can be made double stranded, and cloned as shown in Figure 39.18, and discussed later in Genetic Engineering and Biotechnology 3. Isolation, Sequencing and Synthesis of Genes. However cDNA clones will differ from genomic clones in lacking the introns present in split genes, but may have the advantage of being capable to be expressed in bacteria, which do not have the machinery to process the hnRNA obtained from split genes, into mRNA.
Colony (or plaque) hybridization for screening of libraries
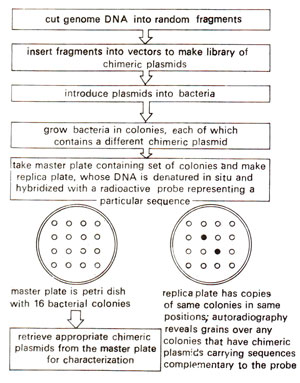 Once a genomic library or cDNA library is available, we may like to use it for isolation of a gene sequence. This can be achieved by colony hybridization technique illustrated in Figure 39.19. In this technique, bacteria carrying chimeric vectors are grown into colonies, which are lysed on nitrocellulose filters. Their DNA is denatured in situ and fixed on the filter, which is hybridized with a radioactively labeled probe carrying a sequence related to the gene to be isolated (usually a cloned cDNA for screening of a genomic library). Colonies carrying this sequence will be identified by dark spots after autoradiography, so that the original chimeric vector carrying the desired gene sequence can be recovered from the original master plate and used for further experiments.
Once a genomic library or cDNA library is available, we may like to use it for isolation of a gene sequence. This can be achieved by colony hybridization technique illustrated in Figure 39.19. In this technique, bacteria carrying chimeric vectors are grown into colonies, which are lysed on nitrocellulose filters. Their DNA is denatured in situ and fixed on the filter, which is hybridized with a radioactively labeled probe carrying a sequence related to the gene to be isolated (usually a cloned cDNA for screening of a genomic library). Colonies carrying this sequence will be identified by dark spots after autoradiography, so that the original chimeric vector carrying the desired gene sequence can be recovered from the original master plate and used for further experiments.
It is possible that a probe may identify more than one clones or that a gene is fragmented in the library. In such a case, one needs to reconstruct the desired sequence using several overlapping sequences available in the library. This is a very routine exercise whenever we like to isolate specific DNA sequences from the genome of a species, or from cDNA derived from mRNA of a specific tissue of a species. Sometimes the library may be available not in the form of bacteria transformed with chimeric DNA molecules, but in the form of chimeric phage particles carrying the cloned segments. In such a situation, a bacterial lawn is infected with a mixture of chimeric phage particles (i.e. the library) and a large number of plaques develop overnight. These plaques can be treated just like the colonies in colony hybridization to identify and isolate the chimeric phage particle carrying the gene of interest. This technique is then described as plaque hybridization.
A cDNA molecule can be made double stranded, and cloned as shown in Figure 39.18, and discussed later in Genetic Engineering and Biotechnology 3. Isolation, Sequencing and Synthesis of Genes. However cDNA clones will differ from genomic clones in lacking the introns present in split genes, but may have the advantage of being capable to be expressed in bacteria, which do not have the machinery to process the hnRNA obtained from split genes, into mRNA.
Colony (or plaque) hybridization for screening of libraries

Fig. 39.19 Technique of construction and screening of a genomic library, using colony hybridization.





