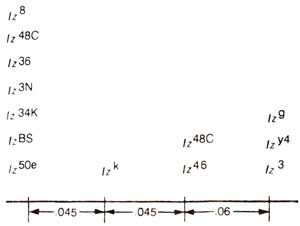
Fig. 14.14. A number of alleles of lozenge occupying four different mutational sites. Recombination frequencies between sites are also given, (redrawn from Strickberger : Genetics).
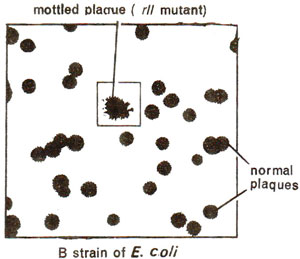
Fig. 14.15. A mottled plaque showing rII mutant among a large number of normal plaques.
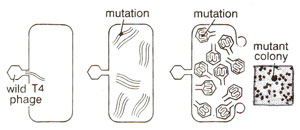
Fig. 14.16. Production of a rII mutant of T4 phage after infection of E. coli by wild T4 phage. (redrawn from Sci. Amer., Jan., 1962).
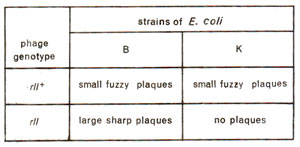
Fig. 14.17. Expression of wild and rII mutant T4phages on two strains (B and K) of E. coli.
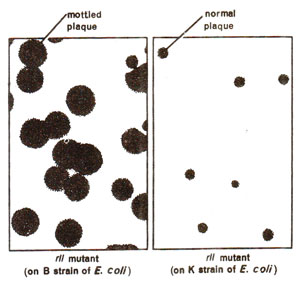
Fig. 14.18. Plaques formed by rII mutant strain on B and K strains of E. coli (normal plaques on K strain are due to wild type T4).
Above explanation makes it clear that a gene expresses itself through a series of steps involved in a sequential synthesis of a product and, therefore, may have one or more functional units. Earlier, we have also shown that there can be several sites in a gene, each capable of being independently involved in mutational and recombinational events. A gene thus is neither a functional, nor a mutational or a recombinational unit, but is a complex locus, whose fine structure should be studied. Such fine structure has been studied in a number of cases using higher resolving power of recombinaton technique. One may like to consider analogies, where a cell is a unit of life, but higher resolution through sophisticated microscopes discovered subcellular particles, and also where an atom is a unit of structure of an element, but subatomic particles like electrons, protons and neutrons are now known.
By corollary, gene is a unit of heredity but its fine structure has been studied through high resolution of the genetic systems. As an electron microscope gives higher resolution for the study of cell and its subcellular structures, a very big population in a genetic experiment will increase resolution of genetic system. Fine structure of a gene was not discovered early in the present century, because limited populations were used for recombination studies, which was unavoidable due to the nature of organisms used for study. When bigger populations were studied, intragenic recombination, whose frequency would be very low, could be discovered and a gene could be resolved into smaller units. This will be illustrated with
the help of few examples.
Fine structure of lozenge locus in Drosophila
The results of
Green and
Green (1949) and
Green (1961) on intragenic recombination in lozenge locus in
Drosophila demonstrated that 14 alleles can be located on atleast four mutational sites separated by distances proportionate to recombination frequencies. These results are presented in Figure 14.14. The alleles on different mutational sites could be subjected to
cis-trans test, and complementation relationships showed that all 14 alleles exhibit lack of complementation among each other, so that they belong to only one functional unit.

Fig. 14.14. A number of alleles of lozenge occupying four different mutational sites. Recombination frequencies between sites are also given, (redrawn from Strickberger : Genetics).

Fig. 14.15. A mottled plaque showing rII mutant among a large number of normal plaques.

Fig. 14.16. Production of a rII mutant of T4 phage after infection of E. coli by wild T4 phage. (redrawn from Sci. Amer., Jan., 1962).

Fig. 14.17. Expression of wild and rII mutant T4phages on two strains (B and K) of E. coli.

Fig. 14.18. Plaques formed by rII mutant strain on B and K strains of E. coli (normal plaques on K strain are due to wild type T4).
Fine structure of rII locus in T4 phage Distinction between wild type and rII mutants using K strain of E. coli.
The most refined analysis of a single gene ever conducted is the one undertaken by
Seymour Benzer for a locus in T4 bacteriophage infecting
E. coli. This locus is known as
rII locus and a mutant at this locus is responsible for the formation of rough plaques or colonies (Figs. 14.15, 14.16). This locus had largest number of rapid lysing (r) mutants, and is called
rII locus. It can be distinguished from other
r loci, by the inability of
rll mutants to produce plaques on lysogenic 'K' strain of
E. coli (Figs. 14.17, 14.18), which carries
X prophage (see
Sexuality and Recombination in Bacteria and Viruses, for lysogeny and prophage). The
rII mutants, may though infect 'K' strain, but can not cause lysis
and are, therefore, unable to produce any plaques
(plaque is a clear space on petri dish formed due to destruction of bacterial cells due to infection by phage and is characteristic of a phage). In contrast, these
rII mutants make large sharp plaques on
E. coli, strain B. The wild type phage T4(
rII+) will make
small and
fuzzy plaques, both on B and K strains (Fig. 14.17). Further, when 'K' was infected simultaneously by
rII+ and
rII, large plaques were formed, since
rII+ helps in lysis so that
rII may express. These distinguishing features enabled Benzer to identify mutants and wild type phages with high efficiency.
Complementation test.
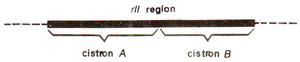
Fig. 14.19. rII region of T4 phage showing two cistrons.
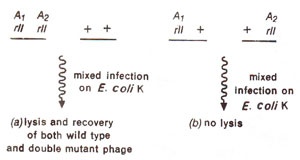
Fig. 14.20. Results of mixed infection by (a) a double mutant (having two rII mutations Ai and A2 belonging to same cistron A) and the wild type strain, phage; (b) two of T4 phage mutant strains, having different rll mutations (Ai and A2) belonging to same cistron A.
Fig. 14.21. Complementation test between mutants to find out if they belong to same or different cistrons. Last row shows cis test to confirm the same relationship, using double mutants (redrawn from Sci. Amer., Jan., 1962).
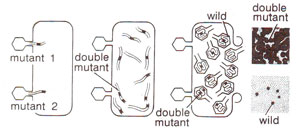
Fig. 14.22. Production of double mutant rII locus due to simultaneous infection of E. coli by two different mutants (redrawn from Sci. Amer., Jan., 1962).
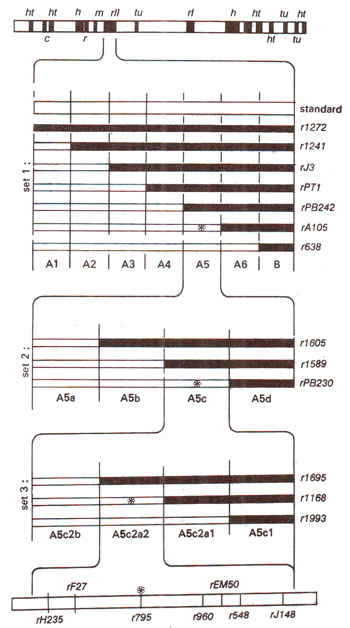
Fig. 14.23. Different overlapping non-revertant (deletion) mutants at rII locus in T4 phage and their use in locating the position of a revertant (point) mutation (marked as *). Note that three sets of overlapping deletion mutations cover narrowing range of areas of rII locus (rII, A5, A5C, A5c2a2) were used.
In order to find out complementation relations between different
rII mutant alleles, Benzer used two different
rII mutants, arbitrarily designated as
rIIx and
rIIy. He allowed mixed infection of K strain by these two mutants. Although in most cases, this does not result into lysis and plaque formation, in some cases it does lead to plaque formation. If two mutants did not form plaques on mixed infection, they were placed in the same group, but if plaques are produced, the two mutants involved in mixed infection were placed in two different groups. In this manner, two groups A and B could be established in
rII region (Fig. 14.19). All mutants with the help of complementation test could be classified in these two groups, in such a manner that two mutants from group A or two mutants from group B could not cause plaque formation but mixed infection by one mutant of group A and another of group B, could cause plaque formation. Since groups A and B are distinguished on the basis of
cis-trans test, these were termed as
cistron A and
cistron B. Mutants belonging to the same cistron i.e. A or B would exibit
cis-trans phenomenon, meaning that they would give wild type only in
cis configuration and not in
trans configuration (Fig. 14.20). Two mutants from different cistrons (A and B) would give wild type (plaque formation) even in
trans configuration, which in other words is called complementation (Fig. 14.21). For the purpose of
cis configuration, a chromosome with two mutants can be obtained through mixed infection followed by recombination (Fig. 14.22).
From the comple-mentation test, it is obvious that in
rII region, two cistrons A and B are independent functionally and must be responsible for sequential synthesis of two separate products, which presumably are polypeptide chains. Therefore, all mutants belonging to one cistron share a common deficiency, which is different from the deficiency due to mutants belonging to the second cistron. When two mutants belong to same cistron, both are deficient for same product and therefore, they can not complement, but when two mutants belong to two different
cistrons, they, being deficient for different products, can complement, and may express wild phenotype
i.e. lysis and plaque formation.
Recombination test.
Once
rII mutants were classified in
cistron A and
cistron B, Benzer was interested in analysing mutants, belonging to same
cistron. It was, therefore, necessary to subject them to recombination test to find out whether they are located on same site or different sites separable by recombination. For this purpose, Benzer classified all mutants into two categories (i)
revertant type- point mutations and (ii)
non-revertant type- deletion mutations (multisite mutations). For recombination analysis, to reduce labour,
Benzer made use of non-revertant mutants which were due to deletions (that these were deletions was obvious from several tests including their non-revertant nature). These deletions were arranged in sets of overlapping deletions representing segments of different sizes in
rII region as shown in Figure 14.23. The principle involved in this technique was that if a particular point mutant lies in the region of a deletion represented by a
rII mutant, then on mixed infection with this deletion mutant, the point mutant will not be able to give rise to a wild type, but if it falls outside the deletion region, it will be able to give wild type recombinant.
Using non-revertant (deletion) mutants having successive overlapping deletions of smaller lengths, one could locate a mutant to a fairly small region. All point mutants located in this particular small segment of
rII region, could, then be subjected to recombination test. For this purpose, two mutants at a time were used for mixed infection on
E. coli B strain and the lysate produced on plaques formed on B strain was used for infection on K strain to find out the frequency of wild type phage particles produced, because this will represent the recombination frequency. By this method, Benzer was successful in estimating recombination frequencies as low as .0001 per cent, although later
Tessman (1965), while studying ultrafine structure of
rII region recorded frequencies as low as .00001 per cent. Thus Benzer was not only able to divide
rII region into
cistrons A and B, but was able to classify mutants belonging to same
cistron into hundreds of mutational sites separable due to recombination.
Through recombination analysis, about 2000 mutations were mapped in
rII region and were found by Benzer to occupy 308 different sites called
mutational sites. These sites were, although scattered fairly evenly throughout the length, some of them were called
hot spots since they had numerous mutations relative to other sites. In contrast, some sites had no mutants and were, therefore, called
zero sites. Benzer eventually estimated 400-500 mutational sites in
rII region and called each of them a unit of mutation or a
muton.

Fig. 14.19. rII region of T4 phage showing two cistrons.

Fig. 14.20. Results of mixed infection by (a) a double mutant (having two rII mutations Ai and A2 belonging to same cistron A) and the wild type strain, phage; (b) two of T4 phage mutant strains, having different rll mutations (Ai and A2) belonging to same cistron A.

Fig. 14.21. Complementation test between mutants to find out if they belong to same or different cistrons. Last row shows cis test to confirm the same relationship, using double mutants (redrawn from Sci. Amer., Jan., 1962).

Fig. 14.22. Production of double mutant rII locus due to simultaneous infection of E. coli by two different mutants (redrawn from Sci. Amer., Jan., 1962).

Fig. 14.23. Different overlapping non-revertant (deletion) mutants at rII locus in T4 phage and their use in locating the position of a revertant (point) mutation (marked as *). Note that three sets of overlapping deletion mutations cover narrowing range of areas of rII locus (rII, A5, A5C, A5c2a2) were used.
























