Termination and antitermination of mRNA synthesis in prokaryotes
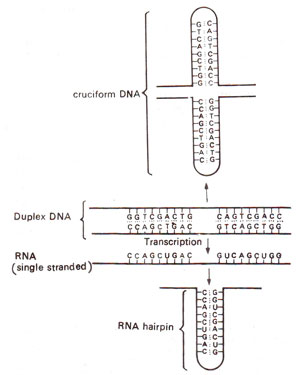
Fig. 32.7. Secondary structures (cruciform DNA and RNA hairpin) formed due to inverted repeats in double stranded DNA and its RNA transcript, (modified from Lewin's "Genes").
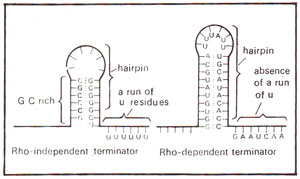
Fig. 32.8. RNA hairpins followed by a run of U residues in rho independent terminator, but not in rho dependent terminator of transcription (modified from Lewin's "Genes").
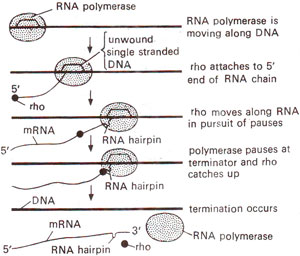
Fig. 32.9. Mechanism of action of rho (p) factor in termination of transcription, involving its travel along RNA till it catches RNA polymerase pausing at a RNA hairpin (modified from Lewin's "Genes").
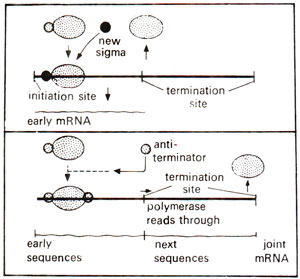
Fig. 32.10. Control of termination by the absence or presence of an antiterminator molecule, which attaches to RNA polymerase enzyme (modified from Lewin's "Gene").
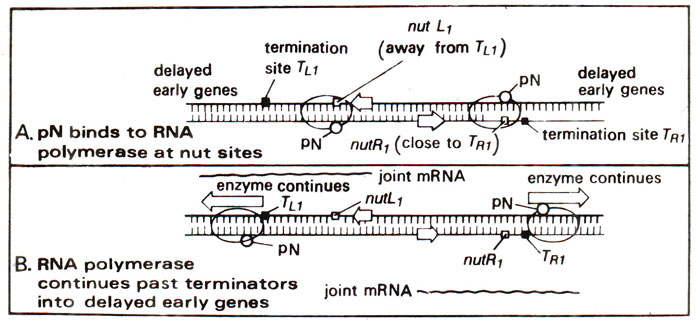
Fig. 32.11. Transcription in phage (λ)showing the role of pN protein. (A) pN protein binds to RNA polymerase. (B) pN protein causes antitermination, i.e. reading through the terminator sites TL1 and Tr1 (modified from Lewin's "Genes").
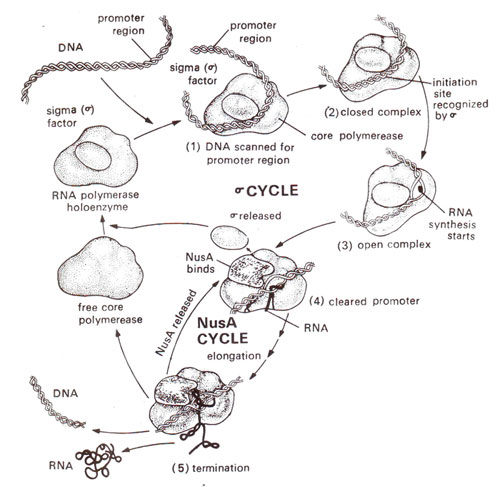
Fig. 32.4. Role of sigma factor Nus A, and core enzyme of RNA polymerase during transcription.
Rho (ρ) dependent and rho (ρ) independent termination of E. coli
The termination signals whenever found on DNA can be of two types : (i) those, which are recognized by a protein factor known as rho (ρ) factor, cause rho (ρ) dependent termination and (ii) those, which do not need rho (ρ) factor for termination cause rho (ρ) independent termination. Rho (ρ) factor is a protein of molecular weight 55,000 and was isolated from E. coli. It causes termination of most mRNA molecules in vitro at relatively low ionic strength, but is not needed for termination of some of the mRNAs. However, in vitro, high ionic strength needs to be maintained to demonstrate rho independent termination. In vivo, distinction between rho dependent and rho independent terminators was possible with the help of mutants deficient for rho factor, but still showing termination at the rho independent sites. In both cases (rho dependent and rho independent terminations), near the termination sites, there is similarity in secondary structure, which is generated, by palindromic sequences like the following :
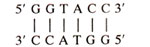
In this palindrome the sequence on both strands is the same when read in 5' to 3' direction. In this palindrome,

also represents an inverse repeat giving dyad symmetry in the sequence. This causes complementarity between bases on same strand (GGT being complementary to ACC read in opposite directions), so that the latter can base pair to form a hairpin in mRNA or cruciform in DNA (Fig. 32.7).
Rho independent terminator differs from rho dependent terminator in the presence of a long run of U residues in mRNA (A residues in DNA) and of a G-C rich region in stem of hairpin structure formed by mRNA (A-T rich structure in DNA) (Fig. 32.8).
Palindromes in mRNA (which is being synthesized) cause formation of hairpins, which cause RNA polymerase to slow down or cause a pause in transcription. The events that follow this pause depend on the type of termination site, (i) At rho independent terminator, due to presence of a long run of U bases in mRNA and A bases in DNA template, there will be very weak rU-dA (RNA-DNA hybrid) base pairs needing very little energy to break. Since RNA polymerase slows down due to hairpin structure, rU-dA bond breaks at any one point causing release of mRNA. (ii) At fho-dependent terminator, rho protein is active as a tetramer. A model for its action proposes that rho (ρ) binds to the 5' end of nascent mRNA chain and moves along the length of mRNA. When RNA polymerase is having a pause due to hairpin structure, rho (ρ) catches up, interacts with RNA polymerase and releases the mRNA molecule (Fig. 32.9), from template accompanied with dissociation.of RNA polymerase and rho (ρ) factor.
Antitermination in phage.
In Regulation of Gene Expression 2. Cricuit of Lytic Cycle and Lysogeny in Bacteriophages, we will show how early, middle and late genes are transcribed in a phage after infection, due to replacement of one sigma factor by another. In such cases new promoters will be needed for each set of genes which are expressed at different times and therefore, need to be recognized by new sigma factors.
There is yet another protein NusA (N utilization substance), which functions like rho (ρ), but is complementary to it. It displaces sigma after initiation and helps in termination (Fig. 32.4). The protein pN perhaps functions by displacing NusA, so that the termination may not take place. (For more details consult "Genes V" by Benjamin Lewin).

Fig. 32.7. Secondary structures (cruciform DNA and RNA hairpin) formed due to inverted repeats in double stranded DNA and its RNA transcript, (modified from Lewin's "Genes").

Fig. 32.8. RNA hairpins followed by a run of U residues in rho independent terminator, but not in rho dependent terminator of transcription (modified from Lewin's "Genes").

Fig. 32.9. Mechanism of action of rho (p) factor in termination of transcription, involving its travel along RNA till it catches RNA polymerase pausing at a RNA hairpin (modified from Lewin's "Genes").

Fig. 32.10. Control of termination by the absence or presence of an antiterminator molecule, which attaches to RNA polymerase enzyme (modified from Lewin's "Gene").

Fig. 32.11. Transcription in phage (λ)showing the role of pN protein. (A) pN protein binds to RNA polymerase. (B) pN protein causes antitermination, i.e. reading through the terminator sites TL1 and Tr1 (modified from Lewin's "Genes").

Fig. 32.4. Role of sigma factor Nus A, and core enzyme of RNA polymerase during transcription.




