Yeast tRNA splicing by cutting and rejoining
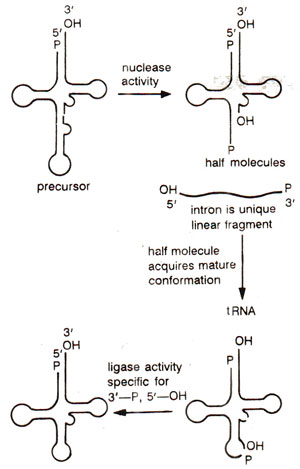
Fig. 33.9. Splicing of yeast tRNA, due to nuclease and ligase activities.
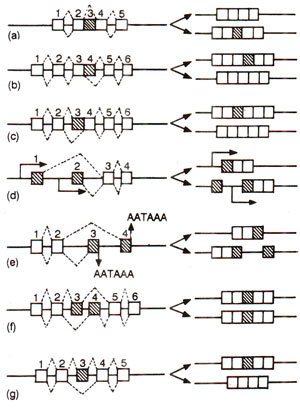
Fig. 30.10. Models of alternative splicing; constitutively spliced exons are shown as white boxes and alternatively spliced exons as hatched boxes; splicing pathways are shown by the diagonal lines; promoters and poly (A) sites are denoted as TATA and AATAAA respectively.
About 40 genes of approximately 400 genes for yeast nuclear tRNAs are interrupted, each with a single intron (14-46 bp), located one nucleotide away from 3' end of anticodon. This intron which may differ in different tRNA genes, has no consensus sequence, but has a sequence complementary to the anticodon. This leads to pairing between anticodon and intron giving a secondary structure, which is recognized by the splicing machinery. The splicing requires following two steps : (i) phosphodiester bond cleavage by an endonuclease; this does not require ATP; and (ii) ligation reaction, which requires ATP and involves bond formation with the help of RNA ligase. the splicing mechanism is illustrated in Figure 33.9.
Trans-splicing of transcripts in chloroplasts and mitochondria
Trans-splicing means that the gene is initially transcribed in pieces and after excision of introns, these pieces join to give rise to mature mRNA. Such trans-splicing reactions involving introns of chloroplast fragmented genes (rps 12 in land plants and psaA in Chlamydomonas reinhardtii)have been reported.
A similar trans-splicing pathway has been reported to generate the mature mRNA for the mitochondrial gene for NAD1 (or NAD5) protein in several plants. In all these cases, coding region of the gene is found scattered among other genes and each coding segment is flanked by intron-Iike sequences. In these trans-splicing reactions, factors (protein as well as RNA) coded by nuclear as well as organellar genes take part.
In the earlier sections of this section, we have discussed the various mechanisms involved in RNA splicing. Generally, the pre-mRNAs undergo processing, so that the non-coding intervening sequences, i.e. introns, are excised, and exons correctly ligated. When this splicing takes place in all cells, without any variation, it is described as 'constitutive splicing' in contrast to 'alternative splicing', which differs in time and space. It has been shown that expression of a large number of genes is regulated by alternative splicing, which gives rise to protein 'isoforms', sharing extensive regions of identity and varying only in specific families. This mechanism of regulation has been used in organisms extending from viruses to mammals and is an economical method of regulating gene expression at the post-transcriptional level.
Retained introns. In some genes an intron may be spliced out or retained. Examples include fibronectin, PDGFA chain and transposase of 'P' element in Drosophila.
Alternative 5' donor sites (or 3' acceptor sites). Introns of different lengths are spliced out to the availability of 2 of more different donor or acceptor sites (donor or acceptor of OH group during transesterification) at the 5' end and 3' end of introns respectively. Examples include adenovirus Ela, SV40, ultrabithorax and transformer genes of Drosophila, etc.

Fig. 33.9. Splicing of yeast tRNA, due to nuclease and ligase activities.

Fig. 30.10. Models of alternative splicing; constitutively spliced exons are shown as white boxes and alternatively spliced exons as hatched boxes; splicing pathways are shown by the diagonal lines; promoters and poly (A) sites are denoted as TATA and AATAAA respectively.
Mutually exclusive internal exons. In some genes, a pair of exons are present, which are neither spliced together, nor retained together, so that in one state one is spliced out and in the other state, the second exon is spliced out giving two isoforms. Examples include genes for a and β tropomyosin, troponin-T, myosin light chain 1/3 and pyruvatekinase.




