Synthesis of telomeric DNA by telomerase
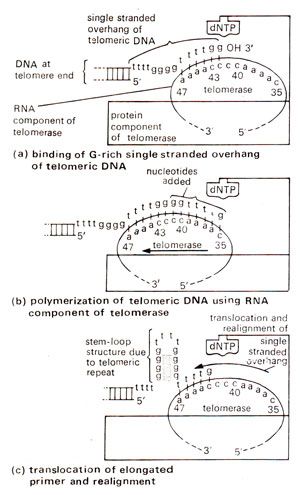
Fig. 26.29. A model for the action of the telomerase from the ciliate Euplotes. This model is based on that first proposed for the Tetrahymcna telomerase, but in the Euplotes telomerase the template nucleotides have been more accurately defined by in vitro studies, (a) the telomeric primer is bound by interactions with the telomerase through recognition of its G:C base-paired secondary structure and Watson-Crick DNA-RNA base pairing between the 3'-nucleotides of the primer with the telomerase RNA template, (b) polymerization copies out to position 35 on the template, (c) the elongated primer translocates backwards, so its new 3' end, becomes aligned with the template, again by base pairing as shown. The unpairing of the RNA-DNA helix in this step is aided by G:G pairing (indicated by shading) within the newly elongated primer. Another cycle of DNA pplymerization occurs, further elongating the primer.
The DNA repeat sequence of telomere has one G-rich strand and the other C rich strand, the G-rich strand having a single stranded overhang. This overhang works as a primer and for its elongation uses as template the RNA component of telomerase enzyme (Fig. 26.29). Telomerase synthesizes only the G-rich strand of telomeres. The complementary C-rich strand is perhaps synthesized (extended) by primase-polymerase mediated discontinuous synthesis, typical of semi-conservative DNA replication, for which extended G-rich strand is used as a template.
For the use of telomerase RNA as template, its sequence should be complementary to telomeric DNA repeat unit, which has been verified. For instance, in Euplotes, 5'CAAAACCCCAAAA3' is found in telomerase, which is used for synthesis of 5'GGGGTTTT3' repeats.

Fig. 26.29. A model for the action of the telomerase from the ciliate Euplotes. This model is based on that first proposed for the Tetrahymcna telomerase, but in the Euplotes telomerase the template nucleotides have been more accurately defined by in vitro studies, (a) the telomeric primer is bound by interactions with the telomerase through recognition of its G:C base-paired secondary structure and Watson-Crick DNA-RNA base pairing between the 3'-nucleotides of the primer with the telomerase RNA template, (b) polymerization copies out to position 35 on the template, (c) the elongated primer translocates backwards, so its new 3' end, becomes aligned with the template, again by base pairing as shown. The unpairing of the RNA-DNA helix in this step is aided by G:G pairing (indicated by shading) within the newly elongated primer. Another cycle of DNA pplymerization occurs, further elongating the primer.




