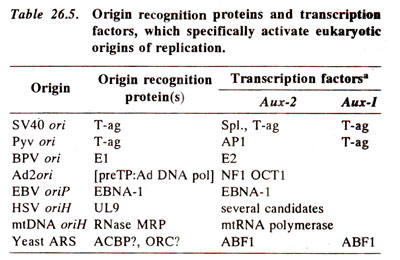
Fig. 26.23. Details of a eukaryotic origin of DNA replication : (a) different components of ori; (b) binding of proteins on different components of ori .
For a study of replication origins, eukaryotes have been classified into (i)
simple genomes including
those from yeast and a number of eukaryotic viruses and (ii)
complex genomes, represented by specific regions of chromosomes belonging to
Xenopus, Chinese Hamster Ovary (CHO) cell lines,
Drosophila and other metazoans. While more detailed information is available for simple genomes, the information for complex genomes is both fragmentary, and difficult to explain. In simple genomes, an origin of replication encompasses 50-1000bp of DNA, but in complex genomes DNA replication occurs at many sites within an
initiation zone, although some sites are highly preferred over others.
In simple genomes,
ori consists of two parts : (i)
core component (analogous to transcription promoter); it consists of an
Origin Recognition Element (
ORE)
, a
DNA-Unwinding Element (
DUE)and an
A/T-rich element, (one strand is A-rich and other strand is T-rich; the A/T rich element is
part of ORE in some cases and it serves as DUE itself in others); (ii) one or more
auxiliary components e.g.
Aux-1 and
Aux-2 (analogous to transcription enhancers). While the sequences of the core component are cis-acting and utilize
origin recognition proteins, the 'auxiliary elements' are trans-acting and provide sites for binding of
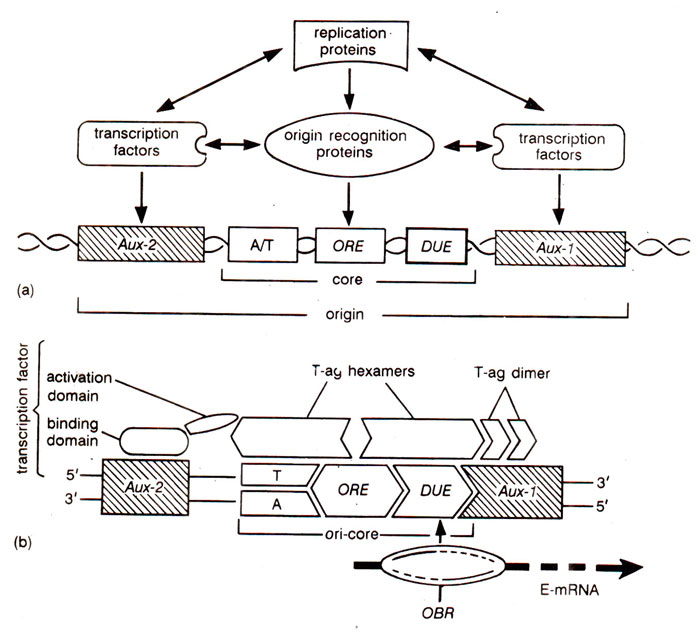
transcription factors (Fig. 26.23; Table 26.5).

Fig. 26.23. Details of a eukaryotic origin of DNA replication : (a) different components of ori; (b) binding of proteins on different components of ori .
Among
simple eukaryotic genomes, replication origins in yeast (
Saccharomyces cerevisiae)were studied in great detail and were termed
'autonomously replicating sequences (
ARSs)
' (Fig. 26.24). It has been shown that these sequences, when present in a plasmid, help high frequency of transformation and allow autonomous replication of plasmids in yeast cells. It has been shown that all DNA replication origins in yeast cells are ARSs, although all ARSs are not replication origins. In all ARSs, meant for DNA replication, there is an 11bp
'AT-rich consensus sequence' or
'ACS' or
'domain A', which is the core component, and is essential but not sufficient for the function of ARS. However, at 3' end of ARS, there is a flanking region called
'domain B' (having three regions, B
1,
B2 and
B3)
, which varies among different ARSs, and is believed to be involved in binding of initiator protein, helix destabilization via DNA unwinding, DNA bending, and binding of accessory protein factors. Therefore, the domain A of ARS functions in concert with three distinct accessory sequences (
B1, B2 and
B3)
, each of which is important but not essential for origin activity. The transcription factor ABF-1 binds at
B3 (analogous to auxiliary components) ' and trans-activates ARS elements, upto lkb away from 'ACS' or 'domain A'.
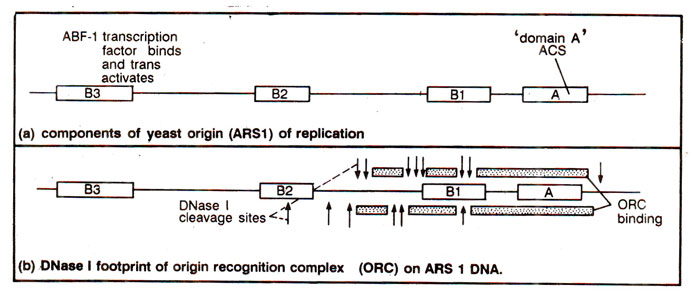
Fig. 26.24. Components of yeast origin of DNA replication, ARS1
A search for initiator factors that recognize ARS1 in a sequence-specific manner, revealed that a complex of six polypeptides binds ACS element of ARS1 with high specificity. This complex is described as
'origin recognition complex (ORC)'. There are also proteins which bind
B3 and
B2 (Fig. 26.24).
The
'origin recognition proteins', listed in Table 26.5, help directly-in DNA unwinding and indirectly in the initiation of replication through their association with 'replication proteins' including DNA polymerases. The transcription factors, which bind the 'auxiliary elements', have each a 'DNA binding domain' and an 'activation domain', the latter involved in activation of 'origin recognition proteins' through protein-protein interaction (Fig. 26.23b). The transcription factors thus facilitate DNA replication through
the following activities : (i) initiation of transcription through
ori core to produce RNA primers for
DNA synthesis; (ii) the assembly of an initiation complex; (iii) activation of initiation complex and (iv) overcoming the repression due to chromatin structure
.
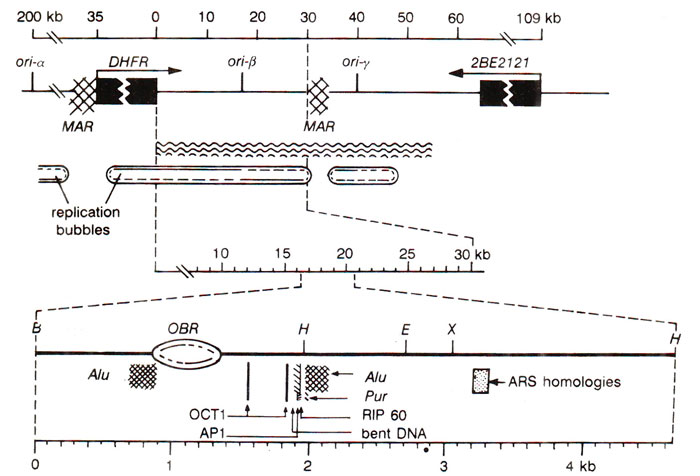
Fig. 26.25. Details of dhfr region (in a CHO cell line) with three initiation zones (ori-a, on'-p, on-y); lower figure shows details of ori-p.
In
complex genomes, a surprising feature is the appearance of
'replication bubbles' (100-500bp) throughout an
'initiation zone' (5000-55000bp). This suggests that replication starts at many sites within an initiation zone. There is, however, evidence that most replication events in metazoan chromosomes begin at sites, which are not randomly chosen, but are specific sites within the larger initiation zone, which is defined by the presence of replication bubbles. There is also evidence that DNA synthesis occurs by the replication fork mechanism and nucleosome segregation is distributive (see
Organization of Genetic Material 1. Packaging of DNA as Nucleosomes in Eukaryotes). The most extensive analysis of mammalian DNA replication (upto 1992) has been carried out in a 300kbp region around the
dhfr gene of Chinese Hamster Ovary (CHO) cells (dhfr = dihydrofolate reductase). In this 300kbp region, replication begins in three different initiation zones referred to as
Ori-α
, Ori-β
, Ori-γ
.
In each of these initiation zones, the origin of replication is described as an
Origin of Bidirectional Replication (OBR)' associated with several elements, which facilitate replication initiation (Fig. 26.25). The
Ori-β, which is the most extensively studied zone in this region is flanked by two '
matrix attachment regions (
MAR)'
. The
OBR within
ori-β is flanked by (i) two
Alu repeats, (ii) sequences associated with DNA amplification, (iii) autonomously replicating plasmid DNA, (iv) a segment of bent DNA, (v) sequences homologous to yeast ARS consensus sequence, (vi) binding sites for atleast four proteins involved in the initiation of DNA replication (RIP60, PUR, OCT1 and AP1; the last two are transcription factors). However, the mechanism involved in the initiation of DNA replication in these multicellular systems, still remains to be fully understood.

Fig. 26.25. Details of dhfr region (in a CHO cell line) with three initiation zones (ori-a, on'-p, on-y); lower figure shows details of ori-p.