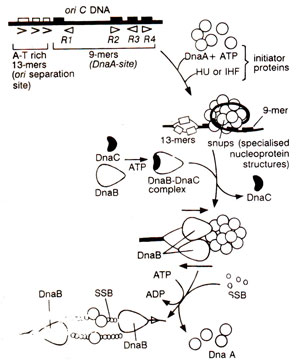
Fig. 26.17. Model of the initiation of DNA replication at oriC of E. coli.
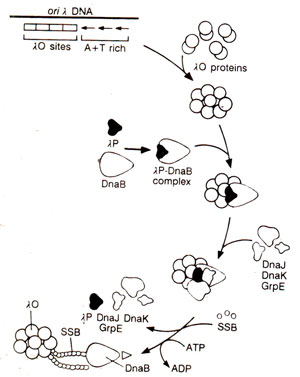
Fig. 26.18. Model of the initiation of DNA replication at oriλof phage λ.
Initiation of replication in double stranded DNA. A comparison of the initiation pathways used at
oriC, oriλ
, oriP1 and
ColE1 (studied
in vitro)suggests three steps : (i) recognition of the origin; (ii) opening of duplex to generate a region of single stranded DNA and (iii) capture of DnaB protein. The events for initiation are sometimes also classified into
'preprinting' (which occurs only at the origin) and
'priming' which recurs with the initiation of each Okazaki fragment during elongation phase. There are variations in different cases, in the mechanism of initiation, and also in the proteins involved in initiation. Only
E. coli oriC will be described here, and differences will be indicated, wherever necessary and possible.
For initiation from
oriC, following steps are involved (as deduced from
in vitro studies :
(i)
DnaA-ATP complex (10-20 monomers of DnaA) binds at 9bp inverted repeat regions (
R1, R2, R3, R4)of
oriC and promotes opening of the DNA in a region of three direct repeats of a 13-bp sequence (called
13-mers; see Fig. 26.17). The opening occurs from right
13-mer leftwards, and requires negatively supercoiled DNA and Hu or
IHF proteins, besides DnaA-ATP complex,
(ii)
DnaB is transferred to exposed single stranded DNA, with the help of
DnaC, in presence of ATP. DnaC, perhaps, recognizes DnaA and thus helps in the transfer of DnaB, but DnaC itself is released from DnaB during the transfer reaction,
(iii)
'DnaB helicase' causes unwinding of the DNA, on addition of ATP,
SSB (single strand binding protein) and
DNA gyrase. This leads to the formation of a
preprinting complex. Unwinding and replication from
oriC proceeds in both directions (bidirectional), SSB binding occurs on single stranded regions, and two DnaB complexes, are loaded one on each strand,
(iv) Unwinding is followed by the synthesis of RNA primers by
DnaG primase (through its interaction with DnaB) and the primers are elongated by
'DNA Pol III HE' (HE = holoenzyme). However,
in vivo, priming of replication, in
E. coli DNA,
X DNA and in the leading DNA strand of Col El, is dependent on synthesis of RNA primer by RNA polymerase. (The mobile complex of helicase and primase has been termed a
'primosome'). On both,
oriC and
oriPl, DnaA and DnaC proteins help in the transfer of DnaB. However in λphage, its own specific proteins λO and λP are available, along with many
E. coli proteins, for replication. λOand λP are analogous to
E. coli's DnaA and DnaC respectively. Activation of DnaB in λphage requires release of
λP, which is assisted by three heat-shock proteins,
DnaK, DnaJ and
GrpE (Fig. 26.18). Loading of DnaB on ColEl is dependent on PriA and Φ X type primosome.

Fig. 26.17. Model of the initiation of DNA replication at oriC of E. coli.

Fig. 26.18. Model of the initiation of DNA replication at oriλof phage λ.
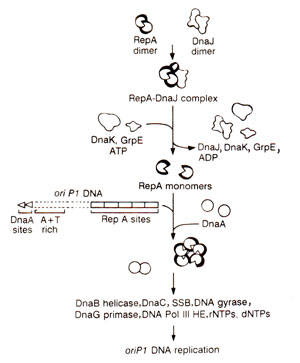
Fig. 26.19. Model of the initiation of DNA replication of oriP1 of phage P1.
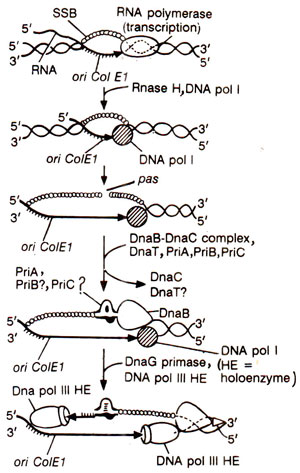
Fig. 26.20. Model of the initiation of DNA replication at ColE1/pBR322 origin.
Replication at
oriP1 of lysogenic plasmid P1 requires the phage-encoded protein,
RepA and many other proteins used by
oriC and
oriλ
. In vitro
oriP1 directed DNA synthesis also depends on three heat shock proteins,
DnaJ, dnaK and
GrpE. DnaJ binds to RepA, and the
RepA-DnaJ complex is recognized by DnaK, which in turn, due to hydrolysis of ATP, triggers the release of active monomeric RepA. The role of GrpE in activation of RepA is not clearly understood. Once active in monomeric form, RepA binds to
oriP1, so that
RepA-oriP1 DNA complex is a substrate for DNA replication. DnaA binds to its sites and leads to unwinding of DNA. DnaB helicase is now transferred to the origin, (perhaps through DnaC-DnaA interaction), and activation of DnaB helicase (without the involvement of DnaJ, Dnak and GrpE) leads to unwinding of the DNA. Now primase synthesizes primers, and
DNA pol III HE catalyzes DNA synthesis (Fig. 26.19).
In vitro, the replication at
oriP1 is unidirectional.
Initiation of DNA replication from ColE1 origin is catalyzed by
E. coli proteins, but differs in several essential features. On the leading strand RNA polymerase synthesizes the transcript forming RNA-DNA hybrid, that is processed by RNase H generating 3' OH group, which serves as a primer for elongation by DNA polymerase I forming DNA-DNA hybrid and displacing the other strand of the plasmid DNA.
The displaced strand exposes a site known as
'primosome assembly site' or pas. A protein-DNA complex, called
' Φ
X-type primosome' (including DnaB) is assembled at this site and is recognized by DnaG primase for initiation of primer synthesis. Thus the primer synthesis in ColEl differs on two strands and the replication is unidirectional. Eventually DNA pol III HE replaces DNA pol I on the leading strand and DnaG primase on the lagging strand for DNA elongation (Fig. 26.20).
Initiation of replication in single stranded DNA phages. In many cases (including phage
M13, which is single stranded and mtDNA) it is the RNA polymerase, which is involved in priming the DNA synthesis through the synthesis of small RNA segment, which precedes DNA synthesis (as earlier discussed). However, in phage G4 and in phage Φ X
174 (both single stranded DNA), in place of RNA polymerase, a much smaller protein called
primase (coded by
dnaG gene) is utilized. This primase enzyme synthesizes RNA from one unique specific site each in phages G4 and M13, but at multiple sites in Φ X 174.
In Φ X 174 (as also in G4 and M13), single stranded DNA is first coated with SSB protein, which is converted to a prepriming intermediate by six proteins named
n, n’, n”, i, DnaB and
DnaC. The prepriming complex of six proteins assembles at a point (hairpin formed due to intrastrand pairing), and dnaG primase can now synthesize short RNA molecules at several sites. The complex of proteins including primase at the priming site is called
primosome. Among the prepriming proteins n
’ seems to be responsible for selecting the site for the prepriming complex and the other proteins have an unknown prepriming function. It has, however, been shown that in the absence of SSB coating, primase in association with DnaB protein can synthesize RNA primer without the need of other five proteins (n, n
’, n
”, i and DnaC), but a prior coating of SSB would need these five proteins for the synthesis of RNA primer.

Fig. 26.19. Model of the initiation of DNA replication of oriP1 of phage P1.

Fig. 26.20. Model of the initiation of DNA replication at ColE1/pBR322 origin.










