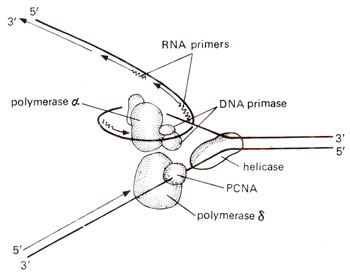
Fig. 26.27. Two different DNA polymerases, synthesizing leading and lagging strands at a eukaryotic DNA replication fork.
During 1987-1989, on the basis of studies on classical enzymology, a proposal was put forward for eukaryotic DNA replication. According to this proposal, unlike DNA replication in prokaryotes, two difference DNA polymerases were required for eukaryotic DNA replication (Fig. 26.27). These two DNA polymerases
(DNA polymerase a and
DNA polymerase δ) were purified and characterized. In 1990, results of a study were reported where DNA replication (using SV40 genome) in a cell free system depended on T antigen (tumour antigen) derived from SV40 and on a number of purified mammalian proteins.
It was demonstrated that DNA polymerase δ synthesizes the DNA on the leading strand (continuous DNA synthesis), whereas DNA polymerase αsynthesizes the DNA on
the lagging strand (discontinuous DNA synthesis). The eight purified components, which may be involved in DNA replication, included the following : (i) T antigen, (ii) replication factor A or
RF-A (also called
RP-A or eukaryotic SSB), (iii)
topoisomerase I, (iv) topoisomerase II, (v) DNA polymerase α
(pol α), which is tightly associated with eukaryotic
primase to form a DNA poly α-primase complex, (vi)
DNA polymerase δ
(pol δ), (vii) proliferating-cell nuclear antigen
(PCNA), also known as
cyclin, (viii) replication factor C or
RF-C. Of these eight components,
T antigen (TAg) was the only component, which was
SV40 encoded. Other components were derived from the host mammalian system.

Fig. 26.27. Two different DNA polymerases, synthesizing leading and lagging strands at a eukaryotic DNA replication fork.
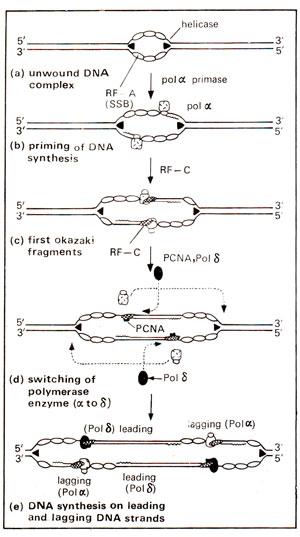
Fig. 26.28. Model for initiating leading and lagging strand synthesis with two DNA polymerases. The first line of the figure shows the unwound complex stage and then four subsequent stages : primer RNA synthesis with primase (priming), first Okazaki fragment synthesis with pol α, switching of DNA polymerases at the 3' end of this first Okazaki fragment, and finally, leading and lagging strand synthesis by pol δ and pol α are indicated (see text for details). Factors involved are DNA helicase (T antigen in the case for SV40), RF-A, pol α-primase, RF-C, PCNA and pol δ. The factors are introduced according to the order of entry into the reaction. Wavy lines are, synthesized primer RNAs, and the dashed lines indicate the introduction of pol δ and the movement of pol α-primase complex along the lagging strand template without dissociation from the replication fork.
TAg has a DNA binding domain and a domain having ATPase and helicase activities. The SV40
ori, involved in the initiation of DNA replication, consists of three defined regions extending over 65 bp : (i) an imperfectly inverted repeat sequence, (ii) a 27 bp binding site for TAg (site II) and (iii) a 17bp AT-rich sequence, meant for DNA unwinding. Adjacent to 65 bp sequence, there is another TAg binding site called site I. These features may be shared by many or all eukaryotic DNA replication origins. Based on the above studies, following steps are suggested for eukaryotic DNA replication, (i) Before DNA synthesis, there is a presynthesis stage of 8-10 minutes duration, for the formation of unwound DNA complex. This stage requires only three purified proteins, namely T antigen (TAg), RF-A and topoisomerase I or II. The T-antigen, using its DNA binding domain, forms a multisubunit complex with site I and site II in the presence of ATP and causes local unwinding. More extensive unwinding follows the association of RF-A and a topoisomerase, with the help of DNA helicase component of TAg. In a number of other viruses also, proteins comparable to TAg are used for the formation of unwound complex for initiation of DNA replication. The topoisomerases help in unwinding of DNA by altering topology of DNA at the replication fork. RF-A or SSB binds to unwound single stranded DNA. (ii) The primase, which is tightly associated with DNA polymerase a, undertakes primer RNA synthesis, (iii) pol αhelps in synthesis of an okazaki fragment in 5' to 3' direction, (iv) RF-C and PCNA (cyclin; see also
Physical Basis of Heredity 3. Genetics, Biochemistry and Dynamics of Cell Division) help in switching of DNA polymerases so that pol α is replaced by pol δ, which then continuously synthesizes DNA on leading strand, (v) Another Okazaki fragment is then synthesized from the replication fork on lagging strand by pol α-primase complex and this step is repeated again and again, till the entire DNA molecule is covered, (vi) The RNA primers are removed and the gaps are filled as in prokaryotic DNA replication. This scheme of DNA replication involving switching, between pol α and pol δ is believed to operate in all eukaryotes (Fig. 26.28).
In another recent study, role of yeast DNA polymerase ε in DNA replication has also been suggested, so that three DNA polymerases (α, δ and ε) are now known to be involved in DNA replication. To accommodate three DNA polymerases functioning at the replication fork, A. Sugino and his colleagues have proposed that polymerase α might function only as an initiation polymerase at both leading and lagging strands (only polymerase a has primase activity), whereas polymerase ε and polymerase δ are involved in elongation of the leading and lagging-strands respectively. There are other models, which suggest the role of either the polymerases α and δ or only polymerase α in eukaryotic DNA replication. It is also possible that in different systems, different enzyme systems are involved.

Fig. 26.28. Model for initiating leading and lagging strand synthesis with two DNA polymerases. The first line of the figure shows the unwound complex stage and then four subsequent stages : primer RNA synthesis with primase (priming), first Okazaki fragment synthesis with pol α, switching of DNA polymerases at the 3' end of this first Okazaki fragment, and finally, leading and lagging strand synthesis by pol δ and pol α are indicated (see text for details). Factors involved are DNA helicase (T antigen in the case for SV40), RF-A, pol α-primase, RF-C, PCNA and pol δ. The factors are introduced according to the order of entry into the reaction. Wavy lines are, synthesized primer RNAs, and the dashed lines indicate the introduction of pol δ and the movement of pol α-primase complex along the lagging strand template without dissociation from the replication fork.








