DNA Coding by Base Sequence
DNA Coding by Base Sequence
Since DNA is the genetic material and is composed of a linear sequence of base pairs, an obvious extension of the Watson-Crick model is that the sequence of base pairs in DNA codes for, and is colinear with, the sequence of amino acids in a protein. The coding hypothesis had to account for the way a string of four different bases—a fourletter alphabet—could dictate the sequence of 20 different amino acids.
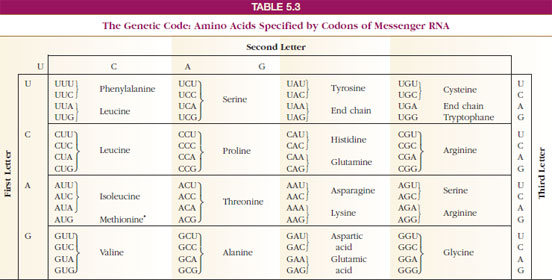
In the coding procedure, obviously there cannot be a 1:1 correlation between four bases and 20 amino acids. If the coding unit (often called a word, or codon) consisted of two bases, only 16 words (42) could be formed, which could not account for 20 amino acids. Therefore the codon had to consist of at least three bases or three letters, because 64 possible words (43) could be formed by four bases when taken as triplets. This means that there could be a considerable redundancy of triplets (codons), since DNA codes for just 20 amino acids. Later work confirmed that nearly all of the amino acids are specified by more than one triplet code (Table 5-3).
DNA shows a surprising stability, both in prokaryotes and in eukaryotes. Interestingly, it is susceptible to damage by harmful chemicals in the environment and radiation. Such damage is usually not permanent, because cells have an efficient repair system. Various types of damage and repair are known, one of which is called excision repair. Ultraviolet irradiation often causes adjacent pyrimidines to link together by covalent bonds (dimerize), preventing transcription and replication. A series of several enzymes “recognizes” the area of the damaged strand and excises the pair of dimerized pyrimidines and several bases following them. DNA polymerase then synthesizes the missing strand along the remaining one, according to the base-pairing rules, and the enzyme DNA ligase joins the end of the new strand to the old one.
Since DNA is the genetic material and is composed of a linear sequence of base pairs, an obvious extension of the Watson-Crick model is that the sequence of base pairs in DNA codes for, and is colinear with, the sequence of amino acids in a protein. The coding hypothesis had to account for the way a string of four different bases—a fourletter alphabet—could dictate the sequence of 20 different amino acids.
In the coding procedure, obviously there cannot be a 1:1 correlation between four bases and 20 amino acids. If the coding unit (often called a word, or codon) consisted of two bases, only 16 words (42) could be formed, which could not account for 20 amino acids. Therefore the codon had to consist of at least three bases or three letters, because 64 possible words (43) could be formed by four bases when taken as triplets. This means that there could be a considerable redundancy of triplets (codons), since DNA codes for just 20 amino acids. Later work confirmed that nearly all of the amino acids are specified by more than one triplet code (Table 5-3).
 |
DNA shows a surprising stability, both in prokaryotes and in eukaryotes. Interestingly, it is susceptible to damage by harmful chemicals in the environment and radiation. Such damage is usually not permanent, because cells have an efficient repair system. Various types of damage and repair are known, one of which is called excision repair. Ultraviolet irradiation often causes adjacent pyrimidines to link together by covalent bonds (dimerize), preventing transcription and replication. A series of several enzymes “recognizes” the area of the damaged strand and excises the pair of dimerized pyrimidines and several bases following them. DNA polymerase then synthesizes the missing strand along the remaining one, according to the base-pairing rules, and the enzyme DNA ligase joins the end of the new strand to the old one.




